mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress
- PMID: 19223440
- PMCID: PMC2661839
- DOI: 10.1261/rna.1403509
mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress
Abstract
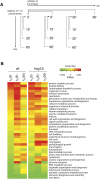
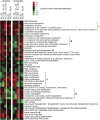
Under stress, cells need to optimize the activity of a wide range of gene products during the response phases: shock, adaptation, and recovery. This requires coordination of several levels of regulation, including turnover and translation efficiencies of mRNAs. Mitogen-activated protein (MAP) kinase pathways are implicated in many aspects of the environmental stress response, including initiation of transcription, translation efficiency, and mRNA turnover. In this study, we analyze mRNA turnover rates and mRNA steady-state levels at different time points following mild hyperosmotic shock in Saccharomyces cerevisiae cells. The regulation of mRNA stability is transient and affects most genes for which there is a change in transcript level. These changes precede and prepare for the changes in steady-state levels, both regarding the initial increase and the later decline of stress-induced mRNAs. The inverse is true for stress-repressed genes, which become stabilized during hyperosmotic stress in preparation of an increase as the cells recover. The MAP kinase Hog1 affects both steady-state levels and stability of stress-responsive transcripts, whereas the Hog1-activated kinase Rck2 influences steady-state levels without a major effect on stability. Regulation of mRNA stability is a wide-spread, but not universal, effect on stress-responsive transcripts during transient hyperosmotic stress. By destabilizing stress-induced mRNAs when their steady-state levels have reached a maximum, the cell prepares for the subsequent recovery phase when these transcripts are to return to normal levels. Conversely, stabilization of stress-repressed mRNAs permits their rapid accumulation in the recovery phase. Our results show that mRNA turnover is coordinated with transcriptional induction.
Figures



References
-
- Albertyn J., Hohmann S., Thevelein J.M., Prior B.A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1994;14:4135–4144. - PMC - PubMed
-
- Arikawa E., Sun Y., Wang J., Zhou Q., Ning B., Dial S.L., Guo L., Yang J. Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study. BMC Genomics. 2008;9:328. doi: 10.1186/1471-2164-9-328. - DOI - PMC - PubMed
-
- Bilsland E., Molin C., Swaminathan S., Ramne A., Sunnerhagen P. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 2004;53:1743–1756. - PubMed
-
- Bilsland E., Hult M., Bell S.D., Sunnerhagen P., Downs J.A. The Bre5/Ubp3 ubiquitin protease complex from budding yeast contributes to the cellular response to DNA damage. DNA Repair (Amst.) 2007;6:1471–1484. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
