RAMI: a tool for identification and characterization of phylogenetic clusters in microbial communities
- PMID: 19223450
- PMCID: PMC2654800
- DOI: 10.1093/bioinformatics/btp051
RAMI: a tool for identification and characterization of phylogenetic clusters in microbial communities
Abstract
Motivation: The most common approach to estimate microbial diversity is based on the analysis of DNA sequences of specific target genes including ribosomal genes. Commonly, the sequences are grouped into operational taxonomic units based on genetic distance (sequence similarity) instead of genetic change (patristic distance). This method may fail to adequately identify clusters of evolutionary related sequences and it provides no information on the phylogenetic structure of the community. An ease-of-use web application for this purpose has been missing.
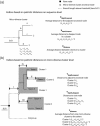
Results: We have developed RAMI, which clusters related nodes in a phylogenetic tree based on the patristic distance. RAMI also produces indices of cluster properties and other indices used in population and community studies on-the-fly.
Availability: RAMI is licensed under GNU GPL and can be run or downloaded from http://www.acgt.se/online.html.
Supplementary information: http://www.acgt.se/RAMI/SuppInfo.
Figures



References
-
- Acinas SG, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. - PubMed
-
- Bohannan BJ, Hughes J. New approaches to analyzing microbial biodiversity data. Curr. Opin. Microbiol. 2003;6:282–287. - PubMed
-
- Cohan FM. Bacterial species and speciation. Syst. Biol. 2001;50:513–524. - PubMed
-
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Seattle: University of Washington; 2005. Department of Genome Sciences.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

