Identification of a cytoplasmic complex that adds a cap onto 5'-monophosphate RNA
- PMID: 19223470
- PMCID: PMC2663312
- DOI: 10.1128/MCB.01325-08
Identification of a cytoplasmic complex that adds a cap onto 5'-monophosphate RNA
Abstract
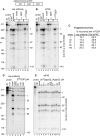
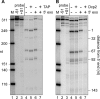
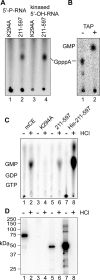
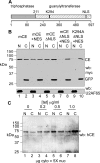
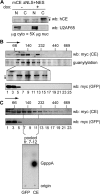
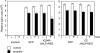
Endonuclease decay of nonsense-containing beta-globin mRNA in erythroid cells generates 5'-truncated products that were reported previously to have a cap or caplike structure. We confirmed that this 5' modification is indistinguishable from the cap on full-length mRNA, and Western blotting, immunoprecipitation, and active-site labeling identified a population of capping enzymes in the cytoplasm of erythroid and nonerythroid cells. Cytoplasmic capping enzyme sediments in a 140-kDa complex that contains a kinase which, together with capping enzyme, converts 5'-monophosphate RNA into 5'-GpppX RNA. Capping enzyme shows diffuse and punctate staining throughout the cytoplasm, and its staining does not overlap with P bodies or stress granules. Expression of inactive capping enzyme in a form that is restricted to the cytoplasm reduced the ability of cells to recover from oxidative stress, thus supporting a role for capping in the cytoplasm and suggesting that some mRNAs may be stored in an uncapped state.
Figures
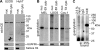


References
-
- Bhattacharyya, S. N., R. Habermacher, U. Martine, E. I. Closs, and W. Filipowicz. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 1251111-1124. - PubMed
-
- Eberle, A. B., S. Lykke-Andersen, O. Muhlemann, and T. H. Jensen. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 1649-55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
