Activation of sterol regulatory element-binding protein by the caspase Drice in Drosophila larvae
- PMID: 19224859
- PMCID: PMC2665088
- DOI: 10.1074/jbc.M900346200
Activation of sterol regulatory element-binding protein by the caspase Drice in Drosophila larvae
Abstract
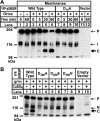
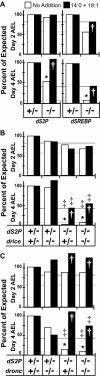
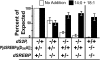
During larval development in Drosophila melanogaster, transcriptional activation of target genes by sterol regulatory element-binding protein (dSREBP) is essential for survival. In all cases studied to date, activation of SREBPs requires sequential proteolysis of the membrane-bound precursor by site-1 protease (S1P) and site-2 protease (S2P). Cleavage by S2P, within the first membrane-spanning helix of SREBP, releases the transcription factor. In contrast to flies lacking dSREBP, flies lacking dS2P are viable. The Drosophila effector caspase Drice cleaves dSREBP, and cleavage requires an Asp residue at position 386, in the cytoplasmic juxtamembrane stalk. The initiator caspase Dronc does not cleave dSREBP, but animals lacking dS2P require both drice and dronc to complete development. They do not require Dcp1, although this effector caspase also can cleave dSREBP in vitro. Cleavage of dSREBP by Drice releases the amino-terminal transcription factor domain of dSREBP to travel to the nucleus where it mediates the increased transcription of target genes needed for lipid synthesis and uptake. Drice-dependent activation of dSREBP explains why flies lacking dS2P are viable, and flies lacking dSREBP itself are not.
Figures




References
-
- Goldstein, J. L., Rawson, R. B., and Brown, M. S. (2002) Arch. Biochem. Biophys. 397 139–148 - PubMed
-
- Srivastava, M., Begovic, E., Chapman, J., Putnam, N. H., Hellsten, U., Kawashima, T., Kuo, A., Mitros, T., Salamov, A., Carpenter, M. L., Signorovitch, A. Y., Moreno, M. A., Kamm, K., Grimwood, J., Schmutz, J., Shapiro, H., Grigoriev, I. V., Buss, L. W., Schierwater, B., Dellaporta, S. L., and Rokhsar, D. S. (2008) Nature 454 955–960 - PubMed
-
- Putnam, N. H., Srivastava, M., Hellsten, U., Dirks, B., Chapman, J., Salamov, A., Terry, A., Shapiro, H., Lindquist, E., Kapitonov, V. V., Jurka, J., Genikhovich, G., Grigoriev, I. V., Lucas, S. M., Steele, R. E., Finnerty, J. R., Technau, U., Martindale, M. Q., and Rokhsar, D. S. (2007) Science 317 86–94 - PubMed
-
- Wang, X., Briggs, M. R., Hua, X., Yokoyama, C., Goldstein, J. L., and Brown, M. S. (1993) J. Biol. Chem. 268 14497–14504 - PubMed
-
- Espenshade, P. J., and Hughes, A. L. (2007) Annu. Rev. Genet. 41 401–427 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

