Laminin receptor involvement in the anti-angiogenic activity of pigment epithelium-derived factor
- PMID: 19224861
- PMCID: PMC2667735
- DOI: 10.1074/jbc.M809259200
Laminin receptor involvement in the anti-angiogenic activity of pigment epithelium-derived factor
Abstract
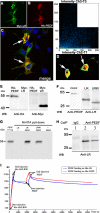
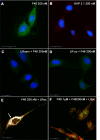
Pigment epithelium-derived factor (PEDF) is a multifunctional protein with neurotrophic, anti-oxidative, and anti-inflammatory properties. It is also one of the most potent endogenous inhibitors of angiogenesis, playing an important role in restricting tumor growth, invasion, and metastasis. Studies show that PEDF binds to cell surface proteins, but little is known about how it exerts its effects. Recently, research identified phospholipase A(2)/nutrin/patatin-like phospholipase domain-containing 2 as one PEDF receptor. To identify other receptors, we performed yeast two-hybrid screening using PEDF as bait and discovered that the non-integrin 37/67-kDa laminin receptor (LR) is another PEDF receptor. Co-immunoprecipitation, His tag pulldown, and surface plasmon resonance assays confirmed the interaction between PEDF and LR. Using the yeast two-hybrid method, we further restricted the LR-interacting domain on PEDF to a 34-amino acid (aa) peptide (aa 44-77) and the PEDF-interacting domain on LR to a 91-aa fragment (aa 120-210). A 25-mer peptide named P46 (aa 46-70), derived from 34-mer, interacts with LR in surface plasmon resonance assays and binds to endothelial cell (EC) membranes. This peptide induces EC apoptosis and inhibits EC migration, tube-like network formation in vitro, and retinal angiogenesis ex vivo, like PEDF. Our results suggest that LR is a real PEDF receptor that mediates PEDF angiogenesis inhibition.
Figures





References
-
- Tombran-Tink, J., Chader, G. G., and Johnson, L. V. (1991) Exp. Eye Res. 53 411–414 - PubMed
-
- Tombran-Tink, J., Mazuruk, K., Rodriguez, I. R., Chung, D., Linker, T., Englander, E., and Chader, G. J. (1996) Mol. Vis. 2 11. - PubMed
-
- Bouck, N. (2002) Trends Mol. Med. 8 330–334 - PubMed
-
- Tombran-Tink, J., and Barnstable, C. J. (2003) Nat. Rev. Neurosci. 4 628–636 - PubMed
-
- Becerra, S. P. (2006) Exp. Eye Res. 82 739–740 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

