Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein
- PMID: 19225007
- PMCID: PMC2668439
- DOI: 10.1128/JVI.02642-08
Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein
Abstract
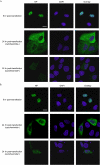
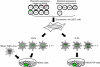
The nucleoprotein (NP), which has multiple functions during the virus life cycle, possesses regions that are highly conserved among influenza A, B, and C viruses. To better understand the roles of highly conserved NP amino acids in viral replication, we conducted a comprehensive mutational analysis. Using reverse genetics, we attempted to generate 74 viruses possessing mutations at conserved amino acids of NP. Of these, 48 mutant viruses were successfully rescued; 26 mutants were not viable, suggesting a critical role of the respective NP amino acids in viral replication. To identify the step(s) in the viral life cycle that is impaired by these NP mutations, we examined viral-genome replication/transcription, NP localization, and incorporation of viral-RNA segments into progeny virions. We identified 15 amino acid substitutions in NP that inhibited viral-genome replication and/or transcription, resulting in significant growth defects of viruses possessing these substitutions. We also found several NP mutations that affected the efficient incorporation of multiple viral-RNA (vRNA) segments into progeny virions even though a single vRNA segment was incorporated efficiently. The respective conserved amino acids in NP may thus be critical for the assembly and/or incorporation of sets of eight vRNA segments.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

