Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes
- PMID: 19226459
- PMCID: PMC2688281
- DOI: 10.1186/gb-2009-10-2-r22
Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes
Erratum in
- Genome Biol. 2012;12(11):411
Abstract
Background: Horizontal transfer (HT) could play an important role in the long-term persistence of transposable elements (TEs) because it provides them with the possibility to avoid the checking effects of host-silencing mechanisms and natural selection, which would eventually drive their elimination from the genome. However, despite the increasing evidence for HT of TEs, its rate of occurrence among the TE pools of model eukaryotic organisms is still unknown.
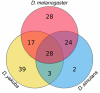
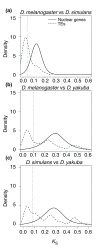
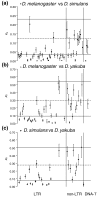
Results: We have extracted and compared the nucleotide sequences of all potentially functional autonomous TEs present in the genomes of Drosophila melanogaster, D. simulans and D. yakuba - 1,436 insertions classified into 141 distinct families - and show that a large fraction of the families found in two or more species display levels of genetic divergence and within-species diversity that are significantly lower than expected by assuming copy-number equilibrium and vertical transmission, and consistent with a recent origin by HT. Long terminal repeat (LTR) retrotransposons form nearly 90% of the HT cases detected. HT footprints are also frequent among DNA transposons (40% of families compared) but rare among non-LTR retroelements (6%). Our results suggest a genomic rate of 0.04 HT events per family per million years between the three species studied, as well as significant variation between major classes of elements.
Conclusions: The genome-wide patterns of sequence diversity of the active autonomous TEs in the genomes of D. melanogaster, D. simulans and D. yakuba suggest that one-third of the TE families originated by recent HT between these species. This result emphasizes the important role of horizontal transmission in the natural history of Drosophila TEs.
Figures

References
-
- Charlesworth B, Sniegowski PD, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. - PubMed
-
- Craig N, Craigie R, Gellert M, Lambowitz A. Mobile DNA II. Washington, DC: ASM Press; 2002.
-
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–412. - PubMed
-
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. - PubMed
-
- Kidwell MG. Transposable elements. In: Gregory TR, editor. The Evolution of the Genome. London: Elsevier Academic Press; 2005. pp. 165–221.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

