Ribonuclease H: the enzymes in eukaryotes
- PMID: 19228196
- PMCID: PMC2746905
- DOI: 10.1111/j.1742-4658.2009.06908.x
Ribonuclease H: the enzymes in eukaryotes
Abstract
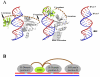
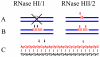
Ribonucleases H are enzymes that cleave the RNA of RNA/DNA hybrids that form during replication and repair and which could lead to DNA instability if they were not processed. There are two main types of RNase H, and at least one of them is present in most organisms. Eukaryotic RNases H are larger and more complex than their prokaryotic counterparts. Eukaryotic RNase H1 has acquired a hybrid binding domain that confers processivity and affinity for the substrate, whereas eukaryotic RNase H2 is composed of three different proteins: the catalytic subunit (2A), similar to the monomeric prokaryotic RNase HII, and two other subunits (2B and 2C) that have no prokaryotic counterparts and as yet unknown functions, but that are necessary for catalysis. In this minireview, we discuss some of the most recent findings on eukaryotic RNases H1 and H2, focusing on the structural data on complexes between human RNase H1 and RNA/DNA hybrids that had provided great detail of how the hybrid binding- and RNase H-domains recognize and cleave the RNA strand of the hybrid substrates. We also describe the progress made in understanding the in vivo function of eukaryotic RNases H. Although prokayotes and some single-cell eukaryotes do not require RNases H for viability, in higher eukaryotes RNases H are essential. Rnaseh1 null mice arrest development around E8.5 because RNase H1 is necessary during embryogenesis for mitochondrial DNA replication. Mutations in any of the three subunits of human RNase H2 cause Aicardi-Goutières syndrome, a human neurological disorder with devastating consequences.
Figures



References
-
- Stein H, Hausen P. Enzyme from Calf Thymus Degrading the RNA Moiety of DNA-RNA Hybrids: Effect on DNA-Dependent RNA Polymerase. Science. 1969;166:393–395. - PubMed
-
- Büsen W, Frank P. Bovine RNases H. In: Crouch RJ, Toulmé JJ, editors. Ribonucleases H. INSERM; Paris: 1998. pp. 113–146.
-
- Crouch RJ, Arudchandran A, Cerritelli SM. RNase H1 of Saccharomyces cerevisiae: methods and nomenclature. Methods Enzymol. 2001;341:395–413. 395-413. - PubMed
-
- Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. Identification of the genes encoding Mn2+-dependent RNase I-III and Mg2+-dependent RNase HIII from Bacillus subtilis: Classification of RNases H into three families. Biochemistry. 1999;38:605–618. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

