UNIQUIMER 3D, a software system for structural DNA nanotechnology design, analysis and evaluation
- PMID: 19228709
- PMCID: PMC2673411
- DOI: 10.1093/nar/gkp005
UNIQUIMER 3D, a software system for structural DNA nanotechnology design, analysis and evaluation
Abstract
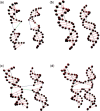
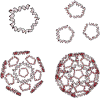
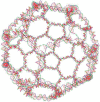
A user-friendly software system, UNIQUIMER 3D, was developed to design DNA structures for nanotechnology applications. It consists of 3D visualization, internal energy minimization, sequence generation and construction of motif array simulations (2D tiles and 3D lattices) functionalities. The system can be used to check structural deformation and design errors under scaled-up conditions. UNIQUIMER 3D has been tested on the design of both existing motifs (holiday junction, 4 x 4 tile, double crossover, DNA tetrahedron, DNA cube, etc.) and nonexisting motifs (soccer ball). The results demonstrated UNIQUIMER 3D's capability in designing large complex structures. We also designed a de novo sequence generation algorithm. UNIQUIMER 3D was developed for the Windows environment and is provided free of charge to the nonprofit research institutions.
Figures










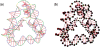
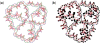

References
-
- Seeman NC. DNA in a material world. Nature. 2003;421:427–431. - PubMed
-
- Seeman NC. DNA engineering and its application to nanotechnology. Trends Biotechnol. 1999;17:437–442. - PubMed
-
- Kallenbach NR, Ma RI, Seeman NC. An immobile nucleic acid junction constructed from oligonucleotides. Nature. 1983;305:829–831.
-
- Li X, Yang X, Qi J, Seeman NC. Antiparallel DNA double crossover molecules as components for nanoconstruction. J. Am. Chem. Soc. 1996;118:6131–6140.
-
- LaBean TH, Yan H, Kopatsch J, Liu F, Winfree E, Reif JH, Seeman NC. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc. 2000;122:1848–1860.

