Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy
- PMID: 19228773
- PMCID: PMC2667287
- DOI: 10.1093/hmg/ddp076
Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy
Abstract
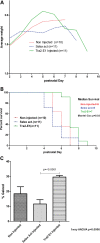
Spinal muscular atrophy (SMA) is a motor neuron disease caused by the loss of survival motor neuron-1 (SMN1). A nearly identical copy gene, SMN2, is present in all SMA patients, which produces low levels of functional protein. Although the SMN2 coding sequence has the potential to produce normal, full-length SMN, approximately 90% of SMN2-derived transcripts are alternatively spliced and encode a truncated protein lacking the final coding exon (exon 7). SMN2, however, is an excellent therapeutic target. Previously, we developed bifunctional RNAs that bound SMN exon 7 and modulated SMN2 splicing. To optimize the efficiency of the bifunctional RNAs, a different antisense target was required. To this end, we genetically verified the identity of a putative intronic repressor and developed bifunctional RNAs that target this sequence. Consequently, there is a 2-fold mechanism of SMN induction: inhibition of the intronic repressor and recruitment of SR proteins via the SR recruitment sequence of the bifunctional RNA. The bifunctional RNAs effectively increased SMN in human primary SMA fibroblasts. Lead candidates were synthesized as 2'-O-methyl RNAs and were directly injected in the central nervous system of SMA mice. Single-RNA injections were able to illicit a robust induction of SMN protein in the brain and throughout the spinal column of neonatal SMA mice. In a severe model of SMA, mean life span was extended following the delivery of bifunctional RNAs. This technology has direct implications for the development of an SMA therapy, but also lends itself to a multitude of diseases caused by aberrant pre-mRNA splicing.
Figures





References
-
- Crawford T.O., Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. - PubMed
-
- Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

