GTPase activation of elongation factor EF-Tu by the ribosome during decoding
- PMID: 19229291
- PMCID: PMC2666022
- DOI: 10.1038/emboj.2009.26
GTPase activation of elongation factor EF-Tu by the ribosome during decoding
Abstract
We have used single-particle reconstruction in cryo-electron microscopy to determine a structure of the Thermus thermophilus ribosome in which the ternary complex of elongation factor Tu (EF-Tu), tRNA and guanine nucleotide has been trapped on the ribosome using the antibiotic kirromycin. This represents the state in the decoding process just after codon recognition by tRNA and the resulting GTP hydrolysis by EF-Tu, but before the release of EF-Tu from the ribosome. Progress in sample purification and image processing made it possible to reach a resolution of 6.4 A. Secondary structure elements in tRNA, EF-Tu and the ribosome, and even GDP and kirromycin, could all be visualized directly. The structure reveals a complex conformational rearrangement of the tRNA in the A/T state and the interactions with the functionally important switch regions of EF-Tu crucial to GTP hydrolysis. Thus, the structure provides insights into the molecular mechanism of signalling codon recognition from the decoding centre of the 30S subunit to the GTPase centre of EF-Tu.
Figures

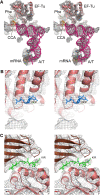
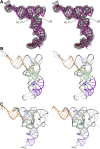

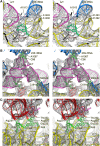

Comment in
-
Long-range signalling in activation of the translational GTPase EF-Tu.EMBO J. 2009 Mar 18;28(6):619-20. doi: 10.1038/emboj.2009.50. EMBO J. 2009. PMID: 19295500 Free PMC article. No abstract available.
References
-
- Abel K, Yoder MD, Hilgenfeld R, Jurnak F (1996) An alpha to beta conformational switch in EF-Tu. Structure 4: 1153–1159 - PubMed
-
- Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF (2006) Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell 23: 865–874 - PubMed
-
- Andersen GR, Nissen P, Nyborg J (2003) Elongation factors in protein biosynthesis. Trends Biochem Sci 28: 434–441 - PubMed
-
- Berchtold H, Reshetnikova L, Reiser CO, Schirmer NK, Sprinzl M, Hilgenfeld R (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365: 126–132 - PubMed
-
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD (2004) tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

