Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3
- PMID: 19229298
- PMCID: PMC2647772
- DOI: 10.1038/emboj.2009.29
Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3
Abstract
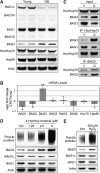
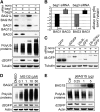
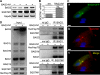
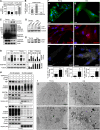
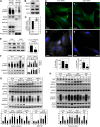
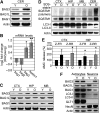
The Hsc/Hsp70 co-chaperones of the BAG (Bcl-2-associated athanogene) protein family are modulators of protein quality control. We examined the specific roles of BAG1 and BAG3 in protein degradation during the aging process. We show that BAG1 and BAG3 regulate proteasomal and macroautophagic pathways, respectively, for the degradation of polyubiquitinated proteins. Moreover, using models of cellular aging, we find that a switch from BAG1 to BAG3 determines that aged cells use more intensively the macroautophagic system for turnover of polyubiquitinated proteins. This increased macroautophagic flux is regulated by BAG3 in concert with the ubiquitin-binding protein p62/SQSTM1. The BAG3/BAG1 ratio is also elevated in neurons during aging of the rodent brain, where, consistent with a higher macroautophagy activity, we find increased levels of the autophagosomal marker LC3-II as well as a higher cathepsin activity. We conclude that the BAG3-mediated recruitment of the macroautophagy pathway is an important adaptation of the protein quality control system to maintain protein homeostasis in the presence of an enhanced pro-oxidant and aggregation-prone milieu characteristic of aging.
Figures

References
-
- Balaban RS, Nemoto S, Finkel T (2005) Mitochondria, oxidants, and aging. Cell 120: 483–495 - PubMed
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319: 916–919 - PubMed
-
- Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292: 1552–1555 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

