Active promoters and insulators are marked by the centrosomal protein 190
- PMID: 19229299
- PMCID: PMC2670862
- DOI: 10.1038/emboj.2009.34
Active promoters and insulators are marked by the centrosomal protein 190
Abstract
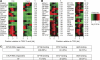
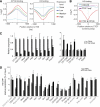
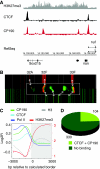
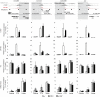
For the compact Drosophila genome, several factors mediating insulator function, such as su(Hw) and dCTCF, have been identified. Recent analyses showed that both these insulator-binding factors are functionally dependent on the same cofactor, CP190. Here we analysed genome-wide binding of CP190 and dCTCF. CP190 binding was detected at CTCF, su(Hw) and GAF sites and unexpectedly at the transcriptional start sites of actively transcribed genes. Both insulator and transcription start site CP190-binding elements are strictly marked by a depletion of histone H3 and, therefore, a loss of nucleosome occupancy. In addition, CP190/dCTCF double occupancy was seen at the borders of many H3K27me3 'islands'. As before, these sites were also depleted of H3. Loss of either dCTCF or CP190 causes an increase of H3 and H3K27 trimethylation at these sites. Thus, for both types of cis-regulatory elements, domain borders and promoters, the chromatin structure is dependent on CP190.
Figures


References
-
- Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

