Allosteric communication occurs via networks of tertiary and quaternary motions in proteins
- PMID: 19229311
- PMCID: PMC2634971
- DOI: 10.1371/journal.pcbi.1000293
Allosteric communication occurs via networks of tertiary and quaternary motions in proteins
Abstract
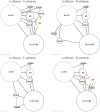
Allosteric proteins bind an effector molecule at one site resulting in a functional change at a second site. We hypothesize that allosteric communication in proteins relies upon networks of quaternary (collective, rigid-body) and tertiary (residue-residue contact) motions. We argue that cyclic topology of these networks is necessary for allosteric communication. An automated algorithm identifies rigid bodies from the displacement between the inactive and the active structures and constructs "quaternary networks" from these rigid bodies and the substrate and effector ligands. We then integrate quaternary networks with a coarse-grained representation of contact rearrangements to form "global communication networks" (GCNs). The GCN reveals allosteric communication among all substrate and effector sites in 15 of 18 multidomain and multimeric proteins, while tertiary and quaternary networks exhibit such communication in only 4 and 3 of these proteins, respectively. Furthermore, in 7 of the 15 proteins connected by the GCN, 50% or more of the substrate-effector paths via the GCN are "interdependent" paths that do not exist via either the tertiary or the quaternary network. Substrate-effector "pathways" typically are not linear but rather consist of polycyclic networks of rigid bodies and clusters of rearranging residue contacts. These results argue for broad applicability of allosteric communication based on structural changes and demonstrate the utility of the GCN. Global communication networks may inform a variety of experiments on allosteric proteins as well as the design of allostery into non-allosteric proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


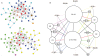

Similar articles
-
Contact rearrangements form coupled networks from local motions in allosteric proteins.Proteins. 2008 Apr;71(1):455-66. doi: 10.1002/prot.21800. Proteins. 2008. PMID: 17957766 Free PMC article.
-
Computational Analysis of Residue Interaction Networks and Coevolutionary Relationships in the Hsp70 Chaperones: A Community-Hopping Model of Allosteric Regulation and Communication.PLoS Comput Biol. 2017 Jan 17;13(1):e1005299. doi: 10.1371/journal.pcbi.1005299. eCollection 2017 Jan. PLoS Comput Biol. 2017. PMID: 28095400 Free PMC article.
-
Motions of Allosteric and Orthosteric Ligand-Binding Sites in Proteins are Highly Correlated.J Chem Inf Model. 2016 Sep 26;56(9):1725-33. doi: 10.1021/acs.jcim.6b00039. Epub 2016 Sep 12. J Chem Inf Model. 2016. PMID: 27580047
-
The Simple and Unique Allosteric Machinery of Thermus caldophilus Lactate Dehydrogenase : Structure-Function Relationship in Bacterial Allosteric LDHs.Adv Exp Med Biol. 2017;925:117-145. doi: 10.1007/5584_2016_171. Adv Exp Med Biol. 2017. PMID: 27815924 Review.
-
Controlling Allosteric Networks in Proteins.Chem Rev. 2016 Jun 8;116(11):6463-87. doi: 10.1021/acs.chemrev.5b00544. Epub 2016 Feb 19. Chem Rev. 2016. PMID: 26894745 Review.
Cited by
-
Dissecting Dynamic Allosteric Pathways Using Chemically Related Small-Molecule Activators.Structure. 2016 Jul 6;24(7):1155-66. doi: 10.1016/j.str.2016.04.010. Epub 2016 May 26. Structure. 2016. PMID: 27238967 Free PMC article.
-
Study, by use of coarse-grained models, of the functionally crucial residues and allosteric pathway of anesthetic regulation of the Gloeobacter violaceus ligand-gated ion channel.Eur Biophys J. 2014 Dec;43(12):623-30. doi: 10.1007/s00249-014-0992-7. Epub 2014 Nov 4. Eur Biophys J. 2014. PMID: 25367560
-
Quantifying Correlations Between Allosteric Sites in Thermodynamic Ensembles.J Chem Theory Comput. 2009 Sep 8;5(9):2486-2502. doi: 10.1021/ct9001812. J Chem Theory Comput. 2009. PMID: 20161451 Free PMC article.
-
A parameterized two-domain thermodynamic model explains diverse mutational effects on protein allostery.Elife. 2024 Jun 5;12:RP92262. doi: 10.7554/eLife.92262. Elife. 2024. PMID: 38836839 Free PMC article.
-
Modulation of Allostery with Multiple Mechanisms by Hotspot Mutations in TetR.J Am Chem Soc. 2024 Jan 31;146(4):2757-2768. doi: 10.1021/jacs.3c12494. Epub 2024 Jan 17. J Am Chem Soc. 2024. PMID: 38231868 Free PMC article.
References
-
- Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. - PubMed
-
- Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. - PubMed
-
- Luque I, Leavitt SA, Freire E. The linkage between protein folding and functional cooperativity: two sides of the same coin? Annu Rev Biophys Biomol Struct. 2002;31:235–256. - PubMed
-
- Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

