Ploidy reductions in murine fusion-derived hepatocytes
- PMID: 19229314
- PMCID: PMC2636893
- DOI: 10.1371/journal.pgen.1000385
Ploidy reductions in murine fusion-derived hepatocytes
Abstract
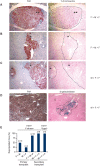
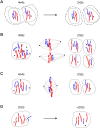
We previously showed that fusion between hepatocytes lacking a crucial liver enzyme, fumarylacetoacetate hydrolase (FAH), and wild-type blood cells resulted in hepatocyte reprogramming. FAH expression was restored in hybrid hepatocytes and, upon in vivo expansion, ameliorated the effects of FAH deficiency. Here, we show that fusion-derived polyploid hepatocytes can undergo ploidy reductions to generate daughter cells with one-half chromosomal content. Fusion hybrids are, by definition, at least tetraploid. We demonstrate reduction to diploid chromosome content by multiple methods. First, cytogenetic analysis of fusion-derived hepatocytes reveals a population of diploid cells. Secondly, we demonstrate marker segregation using ss-galactosidase and the Y-chromosome. Approximately 2-5% of fusion-derived FAH-positive nodules were negative for one or more markers, as expected during ploidy reduction. Next, using a reporter system in which ss-galactosidase is expressed exclusively in fusion-derived hepatocytes, we identify a subpopulation of diploid cells expressing ss-galactosidase and FAH. Finally, we track marker segregation specifically in fusion-derived hepatocytes with diploid DNA content. Hemizygous markers were lost by >or=50% of Fah-positive cells. Since fusion-derived hepatocytes are minimally tetraploid, the existence of diploid hepatocytes demonstrates that fusion-derived cells can undergo ploidy reduction. Moreover, the high degree of marker loss in diploid daughter cells suggests that chromosomes/markers are lost in a non-random fashion. Thus, we propose that ploidy reductions lead to the generation of genetically diverse daughter cells with about 50% reduction in nuclear content. The generation of such daughter cells increases liver diversity, which may increase the likelihood of oncogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Transplanted bone marrow regenerates liver by cell fusion.Nature. 2003 Apr 24;422(6934):901-4. doi: 10.1038/nature01539. Epub 2003 Mar 30. Nature. 2003. PMID: 12665833
-
Proliferative polyploid cells give rise to tumors via ploidy reduction.Nat Commun. 2021 Jan 28;12(1):646. doi: 10.1038/s41467-021-20916-y. Nat Commun. 2021. PMID: 33510149 Free PMC article.
-
Cell fusion is the principal source of bone-marrow-derived hepatocytes.Nature. 2003 Apr 24;422(6934):897-901. doi: 10.1038/nature01531. Epub 2003 Mar 30. Nature. 2003. PMID: 12665832
-
DNA ploidy and autophagic protein degradation as determinants of hepatocellular growth and survival.Cell Biol Toxicol. 1997 Jul;13(4-5):301-15. doi: 10.1023/a:1007487425047. Cell Biol Toxicol. 1997. PMID: 9298250 Review.
-
DNA content and chromosomal composition of malignant human gliomas.Neurol Clin. 1985 Nov;3(4):769-84. Neurol Clin. 1985. PMID: 3001489 Review.
Cited by
-
Constitutive Cdk2 activity promotes aneuploidy while altering the spindle assembly and tetraploidy checkpoints.J Cell Sci. 2013 Mar 1;126(Pt 5):1207-17. doi: 10.1242/jcs.117382. Epub 2013 Jan 15. J Cell Sci. 2013. PMID: 23321641 Free PMC article.
-
Genetic Dissociation of Glycolysis and the TCA Cycle Affects Neither Normal nor Neoplastic Proliferation.Cancer Res. 2017 Nov 1;77(21):5795-5807. doi: 10.1158/0008-5472.CAN-17-1325. Epub 2017 Sep 7. Cancer Res. 2017. PMID: 28883002 Free PMC article.
-
Hippo Signaling Suppresses Cell Ploidy and Tumorigenesis through Skp2.Cancer Cell. 2017 May 8;31(5):669-684.e7. doi: 10.1016/j.ccell.2017.04.004. Cancer Cell. 2017. PMID: 28486106 Free PMC article.
-
Reprogramming Müller glia via in vivo cell fusion regenerates murine photoreceptors.J Clin Invest. 2016 Aug 1;126(8):3104-16. doi: 10.1172/JCI85193. Epub 2016 Jul 18. J Clin Invest. 2016. PMID: 27427986 Free PMC article.
-
Bone marrow stem-cell therapy for genetic and chronic liver diseases.Hepatol Int. 2014 Apr;8(2):166-78. doi: 10.1007/s12072-013-9499-z. Epub 2014 Jan 3. Hepatol Int. 2014. PMID: 26202499
References
-
- Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, et al. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–901. - PubMed
-
- Willenbring H, Bailey AS, Foster M, Akkari Y, Dorrell C, et al. Myelomonocytic cells are sufficient for therapeutic cell fusion in liver. Nat Med. 2004;10:744–748. - PubMed
-
- Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, et al. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12:266–273. - PubMed
-
- Grompe M, Lindstedt S, al-Dhalimy M, Kennaway NG, Papaconstantinou J, et al. Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1995;10:453–460. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

