Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair
- PMID: 19229323
- PMCID: PMC2638008
- DOI: 10.1371/journal.pgen.1000389
Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair
Abstract
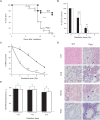
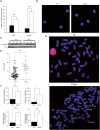
Polymerase micro (Polmicro) is an error-prone, DNA-directed DNA polymerase that participates in non-homologous end-joining (NHEJ) repair. In vivo, Polmicro deficiency results in impaired Vkappa-Jkappa recombination and altered somatic hypermutation and centroblast development. In Polmicro(-/-) mice, hematopoietic development was defective in several peripheral and bone marrow (BM) cell populations, with about a 40% decrease in BM cell number that affected several hematopoietic lineages. Hematopoietic progenitors were reduced both in number and in expansion potential. The observed phenotype correlates with a reduced efficiency in DNA double-strand break (DSB) repair in hematopoietic tissue. Whole-body gamma-irradiation revealed that Polmicro also plays a role in DSB repair in non-hematopoietic tissues. Our results show that Polmicro function is required for physiological hematopoietic development with an important role in maintaining early progenitor cell homeostasis and genetic stability in hematopoietic and non-hematopoietic tissues.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Impairment of the non-homologous end joining and homologous recombination pathways of DNA double strand break repair: Impact on spontaneous and radiation-induced mammary and intestinal tumour risk in Apc min/+ mice.DNA Repair (Amst). 2015 Nov;35:19-26. doi: 10.1016/j.dnarep.2015.08.002. Epub 2015 Sep 6. DNA Repair (Amst). 2015. PMID: 26435054
-
Increased learning and brain long-term potentiation in aged mice lacking DNA polymerase μ.PLoS One. 2013;8(1):e53243. doi: 10.1371/journal.pone.0053243. Epub 2013 Jan 3. PLoS One. 2013. PMID: 23301049 Free PMC article.
-
Hyper-active non-homologous end joining selects for synthetic lethality resistant and pathological Fanconi anemia hematopoietic stem and progenitor cells.Sci Rep. 2016 Feb 26;6:22167. doi: 10.1038/srep22167. Sci Rep. 2016. PMID: 26916217 Free PMC article.
-
Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways.Mutat Res. 2011 Jun 3;711(1-2):61-72. doi: 10.1016/j.mrfmmm.2011.02.005. Epub 2011 Feb 15. Mutat Res. 2011. PMID: 21329706 Review.
-
Templated Insertions: A Smoking Gun for Polymerase Theta-Mediated End Joining.Trends Genet. 2019 Sep;35(9):632-644. doi: 10.1016/j.tig.2019.06.001. Epub 2019 Jul 8. Trends Genet. 2019. PMID: 31296341 Review.
Cited by
-
Exploring the Origin and Physiological Significance of DNA Double Strand Breaks in the Developing Neuroretina.Int J Mol Sci. 2022 Jun 9;23(12):6449. doi: 10.3390/ijms23126449. Int J Mol Sci. 2022. PMID: 35742893 Free PMC article. Review.
-
DNA polymerases and cancer.Nat Rev Cancer. 2011 Feb;11(2):96-110. doi: 10.1038/nrc2998. Nat Rev Cancer. 2011. PMID: 21258395 Free PMC article. Review.
-
Polμ deficiency increases resistance to oxidative damage and delays liver aging.PLoS One. 2014 Apr 1;9(4):e93074. doi: 10.1371/journal.pone.0093074. eCollection 2014. PLoS One. 2014. PMID: 24691161 Free PMC article.
-
DNA expansions generated by human Polμ on iterative sequences.Nucleic Acids Res. 2013 Jan 7;41(1):253-63. doi: 10.1093/nar/gks1054. Epub 2012 Nov 9. Nucleic Acids Res. 2013. PMID: 23143108 Free PMC article.
-
Nonhomologous end joining: a good solution for bad ends.DNA Repair (Amst). 2014 May;17:39-51. doi: 10.1016/j.dnarep.2014.02.008. Epub 2014 Mar 14. DNA Repair (Amst). 2014. PMID: 24630899 Free PMC article. Review.
References
-
- Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair (Amst) 2004;3:827–833. - PubMed
-
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. - PubMed
-
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. - PubMed
-
- Weterings E, van Gent DC. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair (Amst) 2004;3:1425–1435. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

