Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium
- PMID: 19229334
- PMCID: PMC2639726
- DOI: 10.1371/journal.ppat.1000306
Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium
Abstract
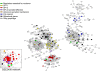
To cause a systemic infection, Salmonella must respond to many environmental cues during mouse infection and express specific subsets of genes in a temporal and spatial manner, but the regulatory pathways are poorly established. To unravel how micro-environmental signals are processed and integrated into coordinated action, we constructed in-frame non-polar deletions of 83 regulators inferred to play a role in Salmonella enteriditis Typhimurium (STM) virulence and tested them in three virulence assays (intraperitoneal [i.p.], and intragastric [i.g.] infection in BALB/c mice, and persistence in 129X1/SvJ mice). Overall, 35 regulators were identified whose absence attenuated virulence in at least one assay, and of those, 14 regulators were required for systemic mouse infection, the most stringent virulence assay. As a first step towards understanding the interplay between a pathogen and its host from a systems biology standpoint, we focused on these 14 genes. Transcriptional profiles were obtained for deletions of each of these 14 regulators grown under four different environmental conditions. These results, as well as publicly available transcriptional profiles, were analyzed using both network inference and cluster analysis algorithms. The analysis predicts a regulatory network in which all 14 regulators control the same set of genes necessary for Salmonella to cause systemic infection. We tested the regulatory model by expressing a subset of the regulators in trans and monitoring transcription of 7 known virulence factors located within Salmonella pathogenicity island 2 (SPI-2). These experiments validated the regulatory model and showed that the response regulator SsrB and the MarR type regulator, SlyA, are the terminal regulators in a cascade that integrates multiple signals. Furthermore, experiments to demonstrate epistatic relationships showed that SsrB can replace SlyA and, in some cases, SlyA can replace SsrB for expression of SPI-2 encoded virulence factors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

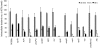


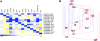

References
-
- Graham SM. Salmonellosis in children in developing and developed countries and populations. Curr Opin Infect Dis. 2002;15:507–512. - PubMed
-
- Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32:263–269. - PubMed
-
- Galan JE, Ginocchio C. The molecular genetic bases of Salmonella entry into mammalian cells. Biochem Soc Trans. 1994;22:301–306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

