Transitional B cells exhibit a B cell receptor-specific nuclear defect in gene transcription
- PMID: 19234182
- PMCID: PMC2694130
- DOI: 10.4049/jimmunol.0802368
Transitional B cells exhibit a B cell receptor-specific nuclear defect in gene transcription
Abstract
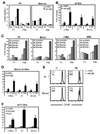
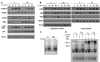
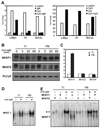
The signaling programs that enforce negative selection in early transitional (T1) B cells in response to BCR engagement remain poorly defined. We conducted a comprehensive comparison of BCR signaling in T1 vs follicular mature splenic B cells. T1, in contrast to follicular mature B cells, failed to express key NF-kappaB target genes in response to BCR engagement and exhibited a striking defect in assembly of an active transcriptional complex at the promoter of the survival and proliferative genes A1 and c-Myc. Surprisingly, and contrary to previous models, classical protein kinase C and IkappaB kinase activation, NF-kappaB nuclear translocation and DNA binding were intact in T1 B cells. Furthermore, despite a marked reduction in NFAT1 expression, differential NFAT or AP-1 activation cannot explain this transcriptional defect. Our combined findings demonstrate that T1 B cells are programmed for signal- and stage-specific "nuclear nonresponsiveness" upon encounter with self-Ags.
Figures






References
-
- Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. - PubMed
-
- Shlomchik MJ. Sites and stages of autoreactive B cell activation and regulation. Immunity. 2008;28:18–28. - PubMed
-
- Hippen KL, Schram BR, Tze LE, Pape KA, Jenkins MK, Behrens TW. In vivo assessment of the relative contributions of deletion, anergy, and editing to B cell self-tolerance. J Immunol. 2005;175:909–916. - PubMed
-
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

