Systems genetics of complex traits in Drosophila melanogaster
- PMID: 19234471
- PMCID: PMC2752214
- DOI: 10.1038/ng.332
Systems genetics of complex traits in Drosophila melanogaster
Abstract
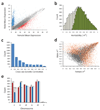
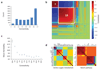
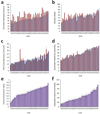
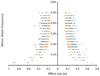
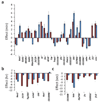
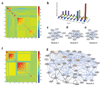
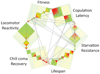
Determining the genetic architecture of complex traits is challenging because phenotypic variation arises from interactions between multiple, environmentally sensitive alleles. We quantified genome-wide transcript abundance and phenotypes for six ecologically relevant traits in D. melanogaster wild-derived inbred lines. We observed 10,096 genetically variable transcripts and high heritabilities for all organismal phenotypes. The transcriptome is highly genetically intercorrelated, forming 241 transcriptional modules. Modules are enriched for transcripts in common pathways, gene ontology categories, tissue-specific expression and transcription factor binding sites. The high degree of transcriptional connectivity allows us to infer genetic networks and the function of predicted genes from annotations of other genes in the network. Regressions of organismal phenotypes on transcript abundance implicate several hundred candidate genes that form modules of biologically meaningful correlated transcripts affecting each phenotype. Overlapping transcripts in modules associated with different traits provide insight into the molecular basis of pleiotropy between complex traits.
Figures
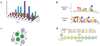
References
-
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Addison Wesley Longman: Harlow; 1996.
-
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, Massachusetts: Sinauer Associates; 1998.
-
- Mackay TFC, Anholt RRH. Of flies and man: Drosophila as a model for human complex traits. Ann. Rev. Genomics Hum. Genetics. 2006;7:339–367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

