Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep
- PMID: 19234472
- PMCID: PMC2683981
- DOI: 10.1038/ng.330
Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep
Abstract
Sleep disorders are common in humans, and sleep loss increases the risk of obesity and diabetes. Studies in Drosophila have revealed molecular pathways and neural tissues regulating sleep; however, genes that maintain genetic variation for sleep in natural populations are unknown. Here, we characterized sleep in 40 wild-derived Drosophila lines and observed abundant genetic variation in sleep architecture. We associated sleep with genome-wide variation in gene expression to identify candidate genes. We independently confirmed that molecular polymorphisms in Catsup (Catecholamines up) are associated with variation in sleep and that P-element mutations in four candidate genes affect sleep and gene expression. Transcripts associated with sleep grouped into biologically plausible genetically correlated transcriptional modules. We confirmed co-regulated gene expression using P-element mutants. Quantitative genetic analysis of natural phenotypic variation is an efficient method for revealing candidate genes and pathways.
Figures
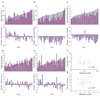

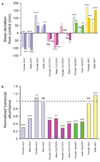

References
-
- Hendricks JC et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. - PubMed
-
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster . Science. 2000;287:1834–1837. - PubMed
-
- Cirelli C et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

