Automated image-based phenotypic analysis in zebrafish embryos
- PMID: 19235725
- PMCID: PMC2861575
- DOI: 10.1002/dvdy.21892
Automated image-based phenotypic analysis in zebrafish embryos
Abstract
Presently, the zebrafish is the only vertebrate model compatible with contemporary paradigms of drug discovery. Zebrafish embryos are amenable to automation necessary for high-throughput chemical screens, and optical transparency makes them potentially suited for image-based screening. However, the lack of tools for automated analysis of complex images presents an obstacle to using the zebrafish as a high-throughput screening model. We have developed an automated system for imaging and analyzing zebrafish embryos in multi-well plates regardless of embryo orientation and without user intervention. Images of fluorescent embryos were acquired on a high-content reader and analyzed using an artificial intelligence-based image analysis method termed Cognition Network Technology (CNT). CNT reliably detected transgenic fluorescent embryos (Tg(fli1:EGFP)(y1)) arrayed in 96-well plates and quantified intersegmental blood vessel development in embryos treated with small molecule inhibitors of anigiogenesis. The results demonstrate it is feasible to adapt image-based high-content screening methodology to measure complex whole organism phenotypes.
(c) 2009 Wiley-Liss, Inc.
Figures

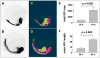


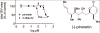

References
-
- Belleri M, Ribatti D, Nicoli S, Cotelli F, Forti L, Vannini V, Stivala LA, Presta M. Antiangiogenic and vascular-targeting activity of the microtubule-destabilizing trans-resveratrol derivative 3,5,4’-trimethoxystilbene. Mol Pharmacol. 2005;67:1451–1459. - PubMed
-
- Belotti D, Vergani V, Drudis T, Borsotti P, Pitelli MR, Viale G, Giavazzi R, Taraboletti G. The microtubule-affecting drug paclitaxel has antiangiogenic activity. Clin Cancer Res. 1996;2:1843–1849. - PubMed
-
- Biberthaler P, Athelogou M, Langer S, Luchting B, Leiderer R, Messmer K. Evaluation of murine liver transmission electron micrographs by an innovative object-based quantitative image analysis system (Cellenger) Eur J Med Res. 2003;8:275–282. - PubMed
-
- Binning G, Baatz M, Klenk J, Schmidt G. Will machines start to think like humans? Europhysics News. 2002;33:45–53.
-
- Brideau C, Gunter B, Pikounis B, Liaw A. Improved statistical methods for hit selection in high-throughput screening. J Biomol Screen. 2003;8:634–647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

