Genome characterization of Pyrenophora tritici-repentis isolates reveals high plasticity and independent chromosomal location of ToxA and ToxB
- PMID: 19236569
- PMCID: PMC6640439
- DOI: 10.1111/j.1364-3703.2008.00520.x
Genome characterization of Pyrenophora tritici-repentis isolates reveals high plasticity and independent chromosomal location of ToxA and ToxB
Abstract
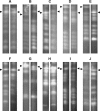
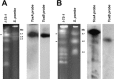
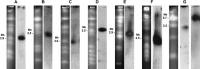
The fungus Pyrenophora tritici-repentis (Died.) causes tan spot, an important leaf disease of wheat worldwide. Isolates of this pathogen have been collected and characterized into eight races on the basis of their ability to produce three different host-selective toxins. The karyotype of 47 isolates was determined by pulsed field gel electrophoresis. The collection originated from different parts of the world and included genotypes from all races. A single isolate was characterized for each of races 3, 4 and 6, whereas fourteen, five, nine, five and eleven isolates were karyotyped for races 1, 2, 5, 7 and 8, respectively. The survey showed that the chromosome number of P. tritici-repentis was highly variable, with some isolates having as few as eight chromosomes, but others having 11 or more. Similarly, the genome size ranged from 25.5 to 48.0 Mb, and individual chromosome sizes ranged from 1.3 to more than 5.7 Mb. Considerable variation was observed in karyotype patterns among the P. tritici-repentis isolates tested. A total of 29 different karyotypes was identified among the 47 isolates. These chromosome level variations were as variable for isolates within a race as for isolates across races. Southern blot analysis of the 47 isolates with ToxA and ToxB probes revealed that the toxin genes were always located on different chromosomes. Furthermore, with six chromosome-specific single-copy probes, the ToxA-carrying chromosome was shown to be homologous among the Ptr ToxA-producing isolates, with a related chromosome in the non-ToxA-producing isolates, suggesting that the chromosome on which ToxA generally resides is of an essential nature. Interestingly, a molecular rearrangement involving a translocation of ToxA to a different chromosome was identified in one isolate.
Figures




References
-
- Anderson, J.A. , Effertz, R.J. , Faris, J.D. , Francl, L.J. , Meinhardt, S.W. and Gill, B.S. (1999) Genetic analysis of sensitivity to Pyrenophora tritici‐repentis necrosis‐inducing toxin in durum and common wheat. Phytopathology, 89, 293–297. - PubMed
-
- Andrie, R.M. , Schoch, C.L. , Hedges, R. , Spatafora, J.W. and Ciuffetti, L.M. (2007) Homologs of ToxB, a host‐selective toxin gene from Pyrenophora tritici‐repentis, are present in the genome of sister‐species Pyrenophora bromi and other members of the Ascomycota. Fungal Genet. Biol. 45, 363–377. - PubMed
-
- Ballance, G.M. , Lamari, L. , Kowatsch, R. and Bernier, C.C. (1996) Cloning expression and occurrence of the gene encoding the Ptr necrosis toxin from Pyrenophora tritici‐repentis . Mol. Plant Pathol . On‐line http://www.bspp.org.uk/mppol/1996/1209ballance.
-
- Bockus, W.W. and Classen, M.M. (1992) Effects of crop rotation and residue management practices on severity of tan spot of winter wheat. Plant Dis. 76, 633–636.
-
- Brasier, C.M. (1987) The dynamics of fungi In: Evolutionary Biology of Fungi (Rayner A.D.M., Brasier C.M. and Moore D., eds), pp. 231–260. Cambridge: Cambridge University Press.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

