Automatic segmentation of the hippocampus and the amygdala driven by hybrid constraints: method and validation
- PMID: 19236922
- PMCID: PMC2677639
- DOI: 10.1016/j.neuroimage.2009.02.013
Automatic segmentation of the hippocampus and the amygdala driven by hybrid constraints: method and validation
Abstract
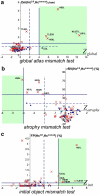
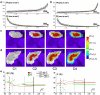
The segmentation from MRI of macroscopically ill-defined and highly variable structures, such as the hippocampus (Hc) and the amygdala (Am), requires the use of specific constraints. Here, we describe and evaluate a fast fully automatic hybrid segmentation that uses knowledge derived from probabilistic atlases and anatomical landmarks, adapted from a semi-automatic method. The algorithm was designed at the outset for application on images from healthy subjects and patients with hippocampal sclerosis. Probabilistic atlases were built from 16 healthy subjects, registered using SPM5. Local mismatch in the atlas registration step was automatically detected and corrected. Quantitative evaluation with respect to manual segmentations was performed on the 16 young subjects, with a leave-one-out strategy, a mixed cohort of 8 controls and 15 patients with epilepsy with variable degrees of hippocampal sclerosis, and 8 healthy subjects acquired on a 3 T scanner. Seven performance indices were computed, among which error on volumes RV and Dice overlap K. The method proved to be fast, robust and accurate. For Hc, results with the new method were: 16 young subjects {RV=5%, K=87%}; mixed cohort {RV=8%, K=84%}; 3 T cohort {RV=9%, K=85%}. Results were better than with atlas-based (thresholded probability map) or semi-automatic segmentations. Atlas mismatch detection and correction proved efficient for the most sclerotic Hc. For Am, results were: 16 young controls {RV=7%, K=85%}; mixed cohort {RV=19%, K=78%}; 3 T cohort {RV=10%, K=77%}. Results were better than with the semi-automatic segmentation, and were also better than atlas-based segmentations for the 16 young subjects.
Figures





References
-
- Ashburner J., Friston K. Unified segmentation. Neuroimage. 2005;26:839–851. - PubMed
-
- Barra V., Boire J.-Y. Automatic segmentation of subcortical brain structures in MR images using information fusion. IEEE Trans. Med. Imag. 2001;20(7):549–568. - PubMed
-
- Barnes J., Foster J., Boyes R.G., Pepple T., Moore E.K., Schott J.M., Frost C. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. Neuroimage. 2008;40:1655–1671. - PubMed
-
- Bloch I., Colliot O., Camara O., Géraud T. Fusion of spatial relationships for guiding recognition, example of brain structure recognition in 3D MRI. Pattern Recogn. Lett. 2005;26:449–457.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

