Placenta-specific methylation of the vitamin D 24-hydroxylase gene: implications for feedback autoregulation of active vitamin D levels at the fetomaternal interface
- PMID: 19237542
- PMCID: PMC2685665
- DOI: 10.1074/jbc.M809542200
Placenta-specific methylation of the vitamin D 24-hydroxylase gene: implications for feedback autoregulation of active vitamin D levels at the fetomaternal interface
Abstract
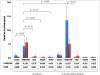
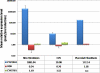
Plasma concentrations of biologically active vitamin D (1,25-(OH)(2)D) are tightly controlled via feedback regulation of renal 1alpha-hydroxylase (CYP27B1; positive) and 24-hydroxylase (CYP24A1; catabolic) enzymes. In pregnancy, this regulation is uncoupled, and 1,25-(OH)(2)D levels are significantly elevated, suggesting a role in pregnancy progression. Epigenetic regulation of CYP27B1 and CYP24A1 has previously been described in cell and animal models, and despite emerging evidence for a critical role of epigenetics in placentation generally, little is known about the regulation of enzymes modulating vitamin D homeostasis at the fetomaternal interface. In this study, we investigated the methylation status of genes regulating vitamin D bioavailability and activity in the placenta. No methylation of the VDR (vitamin D receptor) and CYP27B1 genes was found in any placental tissues. In contrast, the CYP24A1 gene is methylated in human placenta, purified cytotrophoblasts, and primary and cultured chorionic villus sampling tissue. No methylation was detected in any somatic human tissue tested. Methylation was also evident in marmoset and mouse placental tissue. All three genes were hypermethylated in choriocarcinoma cell lines, highlighting the role of vitamin D deregulation in this cancer. Gene expression analysis confirmed a reduced capacity for CYP24A1 induction with promoter methylation in primary cells and in vitro reporter analysis demonstrated that promoter methylation directly down-regulates basal promoter activity and abolishes vitamin D-mediated feedback activation. This study strongly suggests that epigenetic decoupling of vitamin D feedback catabolism plays an important role in maximizing active vitamin D bioavailability at the fetomaternal interface.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

