Chronic enhancement of CREB activity in the hippocampus interferes with the retrieval of spatial information
- PMID: 19237642
- PMCID: PMC2661251
- DOI: 10.1101/lm.1220309
Chronic enhancement of CREB activity in the hippocampus interferes with the retrieval of spatial information
Abstract
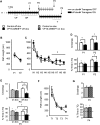
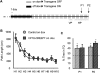
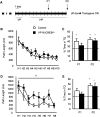
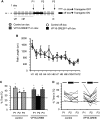
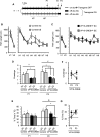
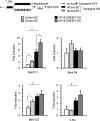
The activation of cAMP-responsive element-binding protein (CREB)-dependent gene expression is thought to be critical for the formation of different types of long-term memory. To explore the consequences of chronic enhancement of CREB function on spatial memory in mammals, we examined spatial navigation in bitransgenic mice that express in a regulated and restricted manner a constitutively active form of CREB, VP16-CREB, in forebrain neurons. We found that chronic enhancement of CREB activity delayed the acquisition of an allocentric strategy to solve the hidden platform task. The ability to turn on and off transgene expression allowed us to dissect the role of CREB in dissociable memory processes. In mice in which transgene expression was turned on during memory acquisition, turning off the transgene re-established the access to the memory trace, whereas in mice in which transgene expression was turned off during acquisition, turning on the transgene impaired memory expression in a reversible manner, indicating that CREB enhancement specifically interfered with the retrieval of spatial information. The defects on spatial navigation in mice with chronic enhancement of CREB function were not corrected by conditions that increased further CREB-dependent activation of hippocampal memory systems, such as housing in an enriched environment. These results along with previous findings in CREB-deficient mutants indicate that the relationship of CREB-mediated plasticity to spatial memory is an inverted-U function, and that optimal learning in the water maze requires accurate regulation of this pathway.
Figures
References
-
- Artola A., von Frijtag J.C., Fermont P.C., Gispen W.H., Schrama L.H., Kamal A., Spruijt B.M. Long-lasting modulation of the induction of LTD and LTP in rat hippocampal CA1 by behavioural stress and environmental enrichment. Eur. J. Neurosci. 2006;23:261–272. - PubMed
-
- Barco A., Alarcon J.M., Kandel E.R. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
