Alternative induction of meiotic recombination from single-base lesions of DNA deaminases
- PMID: 19237686
- PMCID: PMC2674839
- DOI: 10.1534/genetics.109.101683
Alternative induction of meiotic recombination from single-base lesions of DNA deaminases
Abstract
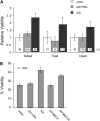
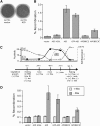
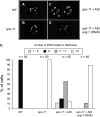
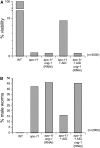
Meiotic recombination enhances genetic diversity as well as ensures proper segregation of homologous chromosomes, requiring Spo11-initiated double-strand breaks (DSBs). DNA deaminases act on regions of single-stranded DNA and deaminate cytosine to uracil (dU). In the immunoglobulin locus, this lesion will initiate point mutations, gene conversion, and DNA recombination. To begin to delineate the effect of induced base lesions on meiosis, we analyzed the effect of expressing DNA deaminases (activation-induced deaminase, AID, and APOBEC3C) in germ cells. We show that meiotic dU:dG lesions can partially rescue a spo11Delta phenotype in yeast and worm. In rec12 Schizosaccharomyces pombe, AID expression increased proper chromosome segregation, thereby enhancing spore viability, and induced low-frequency meiotic crossovers. Expression of AID in the germ cells of Caenorhabditis elegans spo-11 induced meiotic RAD-51 foci formation and chromosomal bivalency and segregation, as well as an increase in viability. RNAi experiments showed that this rescue was dependent on uracil DNA-glycosylase (Ung). Furthermore, unlike ionizing radiation-induced spo-11 rescue, AID expression did not induce large numbers of DSBs during the rescue. This suggests that the products of DNA deamination and base excision repair, such as uracil, an abasic site, or a single-stranded nick, are sufficient to initiate and alter meiotic recombination in uni- and multicellular organisms.
Figures



Comment in
-
Activation-Induced Cytidine Deaminase Does Not Impact Murine Meiotic Recombination.G3 (Bethesda). 2013 Apr 9;3(4):645-655. doi: 10.1534/g3.113.005553. G3 (Bethesda). 2013. PMID: 23550130 Free PMC article.
References
-
- Barreto, V., B. Reina-San-Martin, A. R. Ramiro, K. M. McBride and M. C. Nussenzweig, 2003. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell 12 501–508. - PubMed
-
- Boulton, S. J., A. Gartner, J. Reboul, P. Vaglio, N. Dyson et al., 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295 127–131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

