Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria
- PMID: 19239245
- PMCID: PMC3711378
- DOI: 10.1021/bi8023493
Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria
Abstract
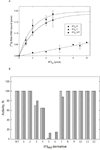
Mitochondrial translational initiation factor 3 (IF3(mt)) is a 29 kDa protein that has N- and C-terminal domains, homologous to prokaryotic IF3, connected by a linker region. The homology domains are preceded and followed by short extensions. No information is currently available on the specific residues in IF3(mt) important for its activity. On the basis of homology models of IF3(mt), mutations were designed in the N-terminal, C-terminal, and linker domains to identify the functionally important regions. Mutation of residues 170-171, and 175 in the C-terminal domain to alanine resulted in a nearly complete loss of activity in initiation complex formation and in the dissociation of mitochondrial 55S ribosomes. However, these mutated proteins bind to the small (28S) subunit of the mammalian mitochondrial ribosome with K(d) values similar to that of the wild-type factor. These mutations appear to lead to a factor defective in the ability to displace the large (39S) subunit of the ribosome from the 55S monosomes in an active process. Other mutations in the N-terminal domain, the linker region, and the C-terminal domain had little or no effect on the ability of IF3(mt) to promote initiation complex formation on mitochondrial 55S ribosomes. Mutation of residues 247 and 248 in the C-terminal extension abolished the ability of IF3(mt) to reduce the level of binding of fMet-tRNA to the ribosome in the absence of mRNA. Our results suggest that IF3(mt) plays an active role in initiation of translation.
Figures






References
-
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. - PubMed
-
- Scheffler I. Mitochondria. New York: Wiley-Liss, Inc; 1999.
-
- Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. - PubMed
-
- Gualerzi C, Pon C. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

