Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement
- PMID: 19239748
- PMCID: PMC2681110
- DOI: 10.3201/eid1503.080631
Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement
Abstract
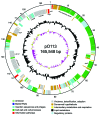
Most Shiga toxin-producing Escherichia coli (STEC) infections that are associated with severe sequelae such as hemolytic uremic syndrome (HUS) are caused by attaching and effacing pathogens that carry the locus of enterocyte effacement (LEE). However, a proportion of STEC isolates that do not carry LEE have been associated with HUS. To clarify the emergence of LEE-negative STEC, we compared the genetic composition of the virulence plasmids pO113 and pO157 from LEE-negative and LEE-positive STEC, respectively. The complete nucleotide sequence of pO113 showed that several plasmid genes were shared by STEC O157:H7. In addition, allelic profiling of the ehxA gene demonstrated that pO113 belongs to a different evolutionary lineage than pO157 and that the virulence plasmids of LEE-negative STEC strains were highly related. In contrast, multilocus sequence typing of 17 LEE-negative STEC isolates showed several clonal groups, suggesting that pathogenic LEE-negative STEC has emerged several times throughout its evolution.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
