The serine protease plasmin cleaves the amino-terminal domain of the NR2A subunit to relieve zinc inhibition of the N-methyl-D-aspartate receptors
- PMID: 19240037
- PMCID: PMC2676017
- DOI: 10.1074/jbc.M805123200
The serine protease plasmin cleaves the amino-terminal domain of the NR2A subunit to relieve zinc inhibition of the N-methyl-D-aspartate receptors
Abstract
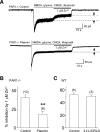
Zinc is hypothesized to be co-released with glutamate at synapses of the central nervous system. Zinc binds to NR1/NR2A N-methyl-d-aspartate (NMDA) receptors with high affinity and inhibits NMDAR function in a voltage-independent manner. The serine protease plasmin can cleave a number of substrates, including protease-activated receptors, and may play an important role in several disorders of the central nervous system, including ischemia and spinal cord injury. Here, we demonstrate that plasmin can cleave the native NR2A amino-terminal domain (NR2A(ATD)), removing the functional high affinity Zn(2+) binding site. Plasmin also cleaves recombinant NR2A(ATD) at lysine 317 (Lys(317)), thereby producing a approximately 40-kDa fragment, consistent with plasmin-induced NR2A cleavage fragments observed in rat brain membrane preparations. A homology model of the NR2A(ATD) predicts that Lys(317) is near the surface of the protein and is accessible to plasmin. Recombinant expression of NR2A with an amino-terminal deletion at Lys(317) is functional and Zn(2+) insensitive. Whole cell voltage-clamp recordings show that Zn(2+) inhibition of agonist-evoked NMDA receptor currents of NR1/NR2A-transfected HEK 293 cells and cultured cortical neurons is significantly reduced by plasmin treatment. Mutating the plasmin cleavage site Lys(317) on NR2A to alanine blocks the effect of plasmin on Zn(2+) inhibition. The relief of Zn(2+) inhibition by plasmin occurs in PAR1(-/-) cortical neurons and thus is independent of interaction with protease-activated receptors. These results suggest that plasmin can directly interact with NMDA receptors, and plasmin may increase NMDA receptor responses through disruption or removal of the amino-terminal domain and relief of Zn(2+) inhibition.
Figures






Similar articles
-
Pharmacological characterization of recombinant NR1/NR2A NMDA receptors with truncated and deleted carboxy termini expressed in Xenopus laevis oocytes.Br J Pharmacol. 2009 Feb;156(3):509-18. doi: 10.1111/j.1476-5381.2008.00040.x. Epub 2009 Jan 16. Br J Pharmacol. 2009. PMID: 19154422 Free PMC article.
-
Zinc inhibition of rat NR1/NR2A N-methyl-D-aspartate receptors.J Physiol. 2008 Feb 1;586(3):763-78. doi: 10.1113/jphysiol.2007.143941. Epub 2007 Nov 29. J Physiol. 2008. PMID: 18048453 Free PMC article.
-
Ethanol inhibition of recombinant NR1/2A receptors: effects of heavy metal chelators and a zinc-insensitive NR2A mutant.Alcohol. 2003 Aug-Oct;31(1-2):71-6. doi: 10.1016/j.alcohol.2003.07.002. Alcohol. 2003. PMID: 14615013
-
Redox modulation of the NMDA receptor.Cell Mol Life Sci. 2000 Oct;57(11):1535-41. doi: 10.1007/pl00000638. Cell Mol Life Sci. 2000. PMID: 11092448 Free PMC article. Review.
-
Amino terminal domain regulation of NMDA receptor function.Eur J Pharmacol. 2004 Oct 1;500(1-3):101-11. doi: 10.1016/j.ejphar.2004.07.015. Eur J Pharmacol. 2004. PMID: 15464024 Review.
Cited by
-
Neuroserpin Differentiates Between Forms of Tissue Type Plasminogen Activator via pH Dependent Deacylation.Front Cell Neurosci. 2016 Jun 15;10:154. doi: 10.3389/fncel.2016.00154. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27378851 Free PMC article.
-
Tailor made plasmin substrates as potential diagnostic tool to test for mastitis.Vet Res Commun. 2014 Dec;38(4):271-7. doi: 10.1007/s11259-014-9611-4. Epub 2014 Jul 6. Vet Res Commun. 2014. PMID: 24997569
-
Cleavage of the NR2B subunit amino terminus of N-methyl-D-aspartate (NMDA) receptor by tissue plasminogen activator: identification of the cleavage site and characterization of ifenprodil and glycine affinities on truncated NMDA receptor.J Biol Chem. 2012 Jul 20;287(30):25520-9. doi: 10.1074/jbc.M112.374397. Epub 2012 May 18. J Biol Chem. 2012. PMID: 22610100 Free PMC article.
-
Impacts of tissue-type plasminogen activator (tPA) on neuronal survival.Front Cell Neurosci. 2015 Oct 16;9:415. doi: 10.3389/fncel.2015.00415. eCollection 2015. Front Cell Neurosci. 2015. PMID: 26528141 Free PMC article. Review.
-
Extracellular proteolysis in structural and functional plasticity of mossy fiber synapses in hippocampus.Front Cell Neurosci. 2015 Nov 4;9:427. doi: 10.3389/fncel.2015.00427. eCollection 2015. Front Cell Neurosci. 2015. PMID: 26582976 Free PMC article. Review.
References
-
- Dingledine, R., Borges, K., Bowie, D., and Traynelis, S. F. (1999) Pharmacol. Rev. 51 7-61 - PubMed
-
- Kemp, J. A., and McKernan, R. M. (2002) Nat. Neurosci. 5 (suppl.) 1039-1042 - PubMed
-
- Stys, P. K., and Lipton, S. A. (2007) Trends Pharmacol. Sci. 28 561-566 - PubMed
-
- Erreger, K., Chen, P. E., Wyllie, D. J., and Traynelis, S. F. (2004) Crit. Rev. Neurobiol. 16 187-224 - PubMed
-
- Paoletti, P., and Neyton, J. (2007) Curr. Opin. Pharmacol. 7 39-47 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

