Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites
- PMID: 19240082
- PMCID: PMC2690485
- DOI: 10.1074/mcp.M800551-MCP200
Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites
Abstract
Small ubiquitin-like modifier (SUMO) is covalently conjugated to its target proteins thereby altering their activity. The mammalian SUMO protein family includes four members (SUMO-1-4) of which SUMO-2 and SUMO-3 are conjugated in a stress-inducible manner. The vast majority of known SUMO substrates are recognized by the single SUMO E2-conjugating enzyme Ubc9 binding to a consensus tetrapeptide (PsiKXE where Psi stands for a large hydrophobic amino acid) or extended motifs that contain phosphorylated or negatively charged amino acids called PDSM (phosphorylation-dependent sumoylation motif) and NDSM (negatively charged amino acid-dependent sumoylation motif), respectively. We identified 382 SUMO-2 targets using a novel method based on SUMO protease treatment that improves separation of SUMO substrates on SDS-PAGE before LC-ESI-MS/MS. We also implemented a software SUMOFI (SUMO motif finder) to facilitate identification of motifs for SUMO substrates from a user-provided set of proteins and to classify the substrates according to the type of SUMO-targeting consensus site. Surprisingly more than half of the substrates lacked any known consensus site, suggesting that numerous SUMO substrates are recognized by a yet unknown consensus site-independent mechanism. Gene ontology analysis revealed that substrates in distinct functional categories display strikingly different prevalences of NDSM sites. Given that different types of motifs are bound by Ubc9 using alternative mechanisms, our data suggest that the preference of SUMO-2 targeting mechanism depends on the biological function of the substrate.
Figures

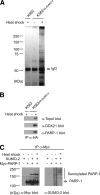
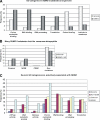
Similar articles
-
In vivo identification of sumoylation sites by a signature tag and cysteine-targeted affinity purification.J Biol Chem. 2010 Jun 18;285(25):19324-9. doi: 10.1074/jbc.M110.106955. Epub 2010 Apr 13. J Biol Chem. 2010. PMID: 20388717 Free PMC article.
-
PDSM, a motif for phosphorylation-dependent SUMO modification.Proc Natl Acad Sci U S A. 2006 Jan 3;103(1):45-50. doi: 10.1073/pnas.0503698102. Epub 2005 Dec 21. Proc Natl Acad Sci U S A. 2006. PMID: 16371476 Free PMC article.
-
In vitro modification of human centromere protein CENP-C fragments by small ubiquitin-like modifier (SUMO) protein: definitive identification of the modification sites by tandem mass spectrometry analysis of the isopeptides.J Biol Chem. 2004 Sep 17;279(38):39653-62. doi: 10.1074/jbc.M405637200. Epub 2004 Jul 22. J Biol Chem. 2004. PMID: 15272016
-
SUMO: getting it on.Biochem Soc Trans. 2007 Dec;35(Pt 6):1409-13. doi: 10.1042/BST0351409. Biochem Soc Trans. 2007. PMID: 18031233 Review.
-
Proteomics strategies to identify SUMO targets and acceptor sites: a survey of RNA-binding proteins SUMOylation.Neuromolecular Med. 2013 Dec;15(4):661-76. doi: 10.1007/s12017-013-8256-8. Epub 2013 Aug 25. Neuromolecular Med. 2013. PMID: 23979992 Review.
Cited by
-
Global SUMO Proteome Responses Guide Gene Regulation, mRNA Biogenesis, and Plant Stress Responses.Front Plant Sci. 2012 Sep 17;3:215. doi: 10.3389/fpls.2012.00215. eCollection 2012. Front Plant Sci. 2012. PMID: 23060889 Free PMC article.
-
Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense.Plant Cell. 2010 Jun;22(6):1998-2016. doi: 10.1105/tpc.109.070961. Epub 2010 Jun 4. Plant Cell. 2010. PMID: 20525853 Free PMC article.
-
Sumoylation of eIF4A2 affects stress granule formation.J Cell Sci. 2016 Jun 15;129(12):2407-15. doi: 10.1242/jcs.184614. Epub 2016 May 9. J Cell Sci. 2016. PMID: 27160682 Free PMC article.
-
Sumoylation in gene regulation, human disease, and therapeutic action.F1000Prime Rep. 2013 Nov 1;5:45. doi: 10.12703/P5-45. eCollection 2013. F1000Prime Rep. 2013. PMID: 24273646 Free PMC article. Review.
-
The serine/arginine-rich protein SF2/ASF regulates protein sumoylation.Proc Natl Acad Sci U S A. 2010 Sep 14;107(37):16119-24. doi: 10.1073/pnas.1004653107. Epub 2010 Aug 30. Proc Natl Acad Sci U S A. 2010. PMID: 20805487 Free PMC article.
References
-
- Geiss-Friedlander, R., and Melchior, F. ( 2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 - PubMed
-
- Nacerddine, K., Lehembre, F., Bhaumik, M., Artus, J., Cohen-Tannoudji, M., Babinet, C., Pandolfi, P. P., and Dejean, A. ( 2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 - PubMed
-
- Lallemand-Breitenbach, V., Jeanne, M., Benhenda, S., Nasr, R., Lei, M., Peres, L., Zhou, J., Zhu, J., Raught, B., and de The, H. ( 2008) Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10, 547–555 - PubMed
-
- Tatham, M. H., Geoffroy, M. C., Shen, L., Plechanovova, A., Hattersley, N., Jaffray, E. G., Palvimo, J. J., and Hay, R. T. ( 2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

