Structure and function of Pseudomonas aeruginosa protein PA1324 (21-170)
- PMID: 19241370
- PMCID: PMC2760366
- DOI: 10.1002/pro.62
Structure and function of Pseudomonas aeruginosa protein PA1324 (21-170)
Abstract
Pseudomonas aeruginosa is the prototypical biofilm-forming gram-negative opportunistic human pathogen. P. aeruginosa is causatively associated with nosocomial infections and with cystic fibrosis. Antibiotic resistance in some strains adds to the inherent difficulties that result from biofilm formation when treating P. aeruginosa infections. Transcriptional profiling studies suggest widespread changes in the proteome during quorum sensing and biofilm development. Many of the proteins found to be upregulated during these processes are poorly characterized from a functional standpoint. Here, we report the solution NMR structure of PA1324, a protein of unknown function identified in these studies, and provide a putative biological functional assignment based on the observed prealbumin-like fold and FAST-NMR ligand screening studies. PA1324 is postulated to be involved in the binding and transport of sugars or polysaccharides associated with the peptidoglycan matrix during biofilm formation.
Figures

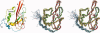



Similar articles
-
Furanone derivatives as quorum-sensing antagonists of Pseudomonas aeruginosa.Appl Microbiol Biotechnol. 2008 Aug;80(1):37-47. doi: 10.1007/s00253-008-1474-6. Epub 2008 Jun 20. Appl Microbiol Biotechnol. 2008. PMID: 18566810
-
PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes.Appl Microbiol Biotechnol. 2008 Feb;78(2):293-307. doi: 10.1007/s00253-007-1308-y. Epub 2007 Dec 22. Appl Microbiol Biotechnol. 2008. PMID: 18157527
-
Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics.Environ Microbiol. 2003 Dec;5(12):1350-69. doi: 10.1046/j.1462-2920.2003.00532.x. Environ Microbiol. 2003. PMID: 14641579
-
Recent perspectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and approaches for treatment and biofilm dispersal.Folia Microbiol (Praha). 2018 Jul;63(4):413-432. doi: 10.1007/s12223-018-0585-4. Epub 2018 Jan 19. Folia Microbiol (Praha). 2018. PMID: 29352409 Review.
-
Application of proteomics to Pseudomonas aeruginosa.Adv Biochem Eng Biotechnol. 2003;83:117-40. doi: 10.1007/3-540-36459-5_5. Adv Biochem Eng Biotechnol. 2003. PMID: 12934928 Review.
Cited by
-
Searching the protein structure database for ligand-binding site similarities using CPASS v.2.BMC Res Notes. 2011 Jan 26;4:17. doi: 10.1186/1756-0500-4-17. BMC Res Notes. 2011. PMID: 21269480 Free PMC article.
-
Cell Envelope Stress Response in Pseudomonas aeruginosa.Adv Exp Med Biol. 2022;1386:147-184. doi: 10.1007/978-3-031-08491-1_6. Adv Exp Med Biol. 2022. PMID: 36258072
-
Identification of genes in the σ²² regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth.mBio. 2012 May 15;3(3):e00094-12. doi: 10.1128/mBio.00094-12. Print 2012. mBio. 2012. PMID: 22589289 Free PMC article.
-
Correlation between protein function and ligand binding profiles.J Proteome Res. 2011 May 6;10(5):2538-45. doi: 10.1021/pr200015d. Epub 2011 Mar 22. J Proteome Res. 2011. PMID: 21366353 Free PMC article.
-
Crystal structure of a putative quorum sensing-regulated protein (PA3611) from the Pseudomonas-specific DUF4146 family.Proteins. 2014 Jun;82(6):1086-92. doi: 10.1002/prot.24455. Epub 2013 Nov 22. Proteins. 2014. PMID: 24174223 Free PMC article.
References
-
- Mirkovic N, Li Z, Parnassa A, Murray D. Strategies for high-throughput comparative modeling: applications to leverage analysis in structural genomics and protein family organization. Proteins. 2007;66:766–777. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

