Conformation of a plasmid replication initiator protein affects its proteolysis by ClpXP system
- PMID: 19241373
- PMCID: PMC2760369
- DOI: 10.1002/pro.68
Conformation of a plasmid replication initiator protein affects its proteolysis by ClpXP system
Abstract
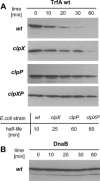
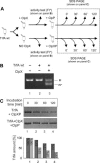
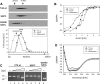
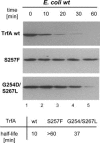
Proteins from the Rep family of DNA replication initiators exist mainly as dimers, but only monomers can initiate DNA replication by interaction with the replication origin (ori). In this study, we investigated both the activation (monomerization) and the degradation of the broad-host-range plasmid RK2 replication initiation protein TrfA, which we found to be a member of a class of DNA replication initiators containing winged helix (WH) domains. Our in vivo and in vitro experiments demonstrated that the ClpX-dependent activation of TrfA leading to replicationally active protein monomers and mutations affecting TrfA dimer formation, result in the inhibition of TrfA protein degradation by the ClpXP proteolytic system. These data revealed that the TrfA monomers and dimers are degraded at substantially different rates. Our data also show that the plasmid replication initiator activity and stability in E. coli cells are affected by ClpXP system only when the protein sustains dimeric form.
Figures



Similar articles
-
The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone.Proc Natl Acad Sci U S A. 1997 Dec 23;94(26):14378-82. doi: 10.1073/pnas.94.26.14378. Proc Natl Acad Sci U S A. 1997. PMID: 9405620 Free PMC article.
-
Opposing effects of DNA on proteolysis of a replication initiator.Nucleic Acids Res. 2012 Feb;40(3):1148-59. doi: 10.1093/nar/gkr813. Epub 2011 Oct 5. Nucleic Acids Res. 2012. PMID: 21976729 Free PMC article.
-
Handcuffing reversal is facilitated by proteases and replication initiator monomers.Nucleic Acids Res. 2017 Apr 20;45(7):3953-3966. doi: 10.1093/nar/gkx166. Nucleic Acids Res. 2017. PMID: 28335002 Free PMC article.
-
[Bacterial ClpX protease structure and function--a review].Wei Sheng Wu Xue Bao. 2010 Oct;50(10):1281-7. Wei Sheng Wu Xue Bao. 2010. PMID: 21141460 Review. Chinese.
-
Remodeling protein complexes: insights from the AAA+ unfoldase ClpX and Mu transposase.Protein Sci. 2005 Aug;14(8):1945-54. doi: 10.1110/ps.051417505. Protein Sci. 2005. PMID: 16046622 Free PMC article. Review.
Cited by
-
Plasmid replication initiator interactions with origin 13-mers and polymerase subunits contribute to strand-specific replisome assembly.Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):E4188-96. doi: 10.1073/pnas.1504926112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195759 Free PMC article.
-
Mechanisms of Theta Plasmid Replication in Enterobacteria and Implications for Adaptation to Its Host.EcoSal Plus. 2020 Nov;9(1):10.1128/ecosalplus.ESP-0026-2019. doi: 10.1128/ecosalplus.ESP-0026-2019. EcoSal Plus. 2020. PMID: 33210586 Free PMC article. Review.
-
Defining a novel domain that provides an essential contribution to site-specific interaction of Rep protein with DNA.Nucleic Acids Res. 2021 Apr 6;49(6):3394-3408. doi: 10.1093/nar/gkab113. Nucleic Acids Res. 2021. PMID: 33660784 Free PMC article.
-
ClpAP protease is a universal factor that activates the parDE toxin-antitoxin system from a broad host range RK2 plasmid.Sci Rep. 2018 Oct 16;8(1):15287. doi: 10.1038/s41598-018-33726-y. Sci Rep. 2018. PMID: 30327496 Free PMC article.
-
Roles of long and short replication initiation proteins in the fate of IncP-1 plasmids.J Bacteriol. 2012 Mar;194(6):1533-43. doi: 10.1128/JB.06395-11. Epub 2012 Jan 6. J Bacteriol. 2012. PMID: 22228734 Free PMC article.
References
-
- Giraldo R. Common domains in the initiators of DNA replication in Bacteria, Archaea, and Eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol Rev. 2003;26:533–554. - PubMed
-
- Cunningham EL, Berger JM. Unraveling the early steps of prokaryotic replication. Curr Opin Struct Biol. 2005;15:68–76. - PubMed
-
- Kornacki JA, West AH, Firshein W. Proteins encoded by the trans-acting replication and maintenance regions of broad host range plasmid RK2. Plasmid. 1984;11:48–57. - PubMed
-
- Shingler V, Thomas CM. Analysis of the trfA region of broad host-range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J Mol Biol. 1984;175:229–249. - PubMed
-
- Perri S, Helinski DR, Toukdarian A. Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host range plasmid RK2. J Biol Chem. 1991;266:12536–12543. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

