The ZNF217 oncogene is a candidate organizer of repressive histone modifiers
- PMID: 19242095
- PMCID: PMC2929765
- DOI: 10.4161/epi.4.2.7953
The ZNF217 oncogene is a candidate organizer of repressive histone modifiers
Abstract
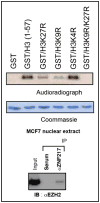
The zinc finger protein 217 (ZNF217) is an important oncogene based on the high frequency of amplification and overexpression in many cancer types, but its molecular mode of gene regulation is poorly understood. We purified a complex of nuclear ZNF217-binding proteins by affinity chromatography and identified its components by mass spectrometry as Jarid1b/Plu-1, G9a, LSD1, CoREST and CtBP1. Individual binding of these with ZNF217 was confirmed by co-immunoprecipiation (IP). Known activities of these proteins suggested a role of the ZNF217 complex in histone modification. Using in vitro assays the following activities were demonstrated: Histone H3 lysine 4 trimethyl (H3K4me3) demethylase activity, which co-fractionated with Jarid1b/Plu-1 in anion-exchange chromatography; H3K9 methylation, the known principal activity of G9a; and H3K27 methylation. The latter suggested EZH2 as another ZNF217 binding candidate, which could be confirmed by co-IP. Taken together, these findings suggest that ZNF217 assembles a distinct set of histone modifying proteins at target DNA sites that act synergistically in transcriptional repression.
Figures



References
-
- Quinlan KG, Verger A, Yaswen P, Crossley M. Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochim Biophys Acta. 2007;1775:333–40. - PubMed
-
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–5. - PubMed
-
- Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J Biol Chem. 2003;278:7234–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
