Adaptation of HIV-1 to human leukocyte antigen class I
- PMID: 19242411
- PMCID: PMC3148020
- DOI: 10.1038/nature07746
Adaptation of HIV-1 to human leukocyte antigen class I
Abstract
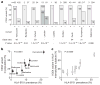
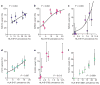
The rapid and extensive spread of the human immunodeficiency virus (HIV) epidemic provides a rare opportunity to witness host-pathogen co-evolution involving humans. A focal point is the interaction between genes encoding human leukocyte antigen (HLA) and those encoding HIV proteins. HLA molecules present fragments (epitopes) of HIV proteins on the surface of infected cells to enable immune recognition and killing by CD8(+) T cells; particular HLA molecules, such as HLA-B*57, HLA-B*27 and HLA-B*51, are more likely to mediate successful control of HIV infection. Mutation within these epitopes can allow viral escape from CD8(+) T-cell recognition. Here we analysed viral sequences and HLA alleles from >2,800 subjects, drawn from 9 distinct study cohorts spanning 5 continents. Initial analysis of the HLA-B*51-restricted epitope, TAFTIPSI (reverse transcriptase residues 128-135), showed a strong correlation between the frequency of the escape mutation I135X and HLA-B*51 prevalence in the 9 study cohorts (P = 0.0001). Extending these analyses to incorporate other well-defined CD8(+) T-cell epitopes, including those restricted by HLA-B*57 and HLA-B*27, showed that the frequency of these epitope variants (n = 14) was consistently correlated with the prevalence of the restricting HLA allele in the different cohorts (together, P < 0.0001), demonstrating strong evidence of HIV adaptation to HLA at a population level. This process of viral adaptation may dismantle the well-established HLA associations with control of HIV infection that are linked to the availability of key epitopes, and highlights the challenge for a vaccine to keep pace with the changing immunological landscape presented by HIV.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- G0500384/MRC_/Medical Research Council/United Kingdom
- R01AI64060/AI/NIAID NIH HHS/United States
- 1 R01 AI067073/AI/NIAID NIH HHS/United States
- G0501777/MRC_/Medical Research Council/United Kingdom
- P30 AI110527/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AI046995/AI/NIAID NIH HHS/United States
- R01 AI067073/AI/NIAID NIH HHS/United States
- R01 AI064060/AI/NIAID NIH HHS/United States
- R01AI46995/AI/NIAID NIH HHS/United States
- R01 AI060460/AI/NIAID NIH HHS/United States
- G108/626/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

