Unique signatures of natural background radiation on human Y chromosomes from Kerala, India
- PMID: 19242544
- PMCID: PMC2644265
- DOI: 10.1371/journal.pone.0004541
Unique signatures of natural background radiation on human Y chromosomes from Kerala, India
Abstract
Background: The most frequently observed major consequences of ionizing radiation are chromosomal lesions and cancers, although the entire genome may be affected. Owing to its haploid status and absence of recombination, the human Y chromosome is an ideal candidate to be assessed for possible genetic alterations induced by ionizing radiation. We studied the human Y chromosome in 390 males from the South Indian state of Kerala, where the level of natural background radiation (NBR) is ten-fold higher than the worldwide average, and that from 790 unexposed males as control.
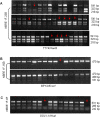
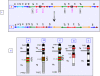
Results: We observed random microdeletions in the Azoospermia factor (AZF) a, b and c regions in >90%, and tandem duplication and copy number polymorphism (CNP) of 11 different Y-linked genes in about 80% of males exposed to NBR. The autosomal homologues of Y-linked CDY genes largely remained unaffected. Multiple polymorphic copies of the Y-linked genes showing single Y-specific signals suggested their tandem duplication. Some exposed males showed unilocus duplication of DAZ genes resulting in six copies. Notably, in the AZFa region, approximately 25% of exposed males showed deletion of the DBY gene, whereas flanking genes USP9Y and UTY remained unaffected. All these alterations were detected in blood samples but not in the germline (sperm) samples.
Conclusions: Exposure to high levels of NBR correlated with several interstitial polymorphisms of the human Y chromosome. CNPs and enhanced transcription of the SRY gene after duplication are envisaged to compensate for the loss of Y chromosome in some cells. The aforesaid changes, confined to peripheral blood lymphocytes, suggest a possible innate mechanism protecting the germline DNA from the NBR. Genome analysis of a larger population focusing on greater numbers of genes may provide new insights into the mechanisms and risks of the resultant genetic damages. The present work demonstrates unique signatures of NBR on human Y chromosomes from Kerala, India.
Conflict of interest statement
Figures








References
-
- Sankaranarayanan K. Ionizing radiations and genetic risks X. The potential “disease phenotypes” of radiation-induced genetic damage in humans: perspectives from human molecular biology and radiation genetics. Mutation Research. 1999;429:45–83. - PubMed
-
- 1993. UNSCEAR United Nations Scientific Committee on the Effects of Atomic Radiations, The 1993 Report to the General Assembly WITH Scientific Annexes, United Nations, New York 1993.
-
- Dubrova YE, Bersimbaev RI, Djansugurova LB, Tankimanova MK, Mamyrbaeva ZZ, et al. Nuclear weapons tests and human germline mutation rate. Science. 2002;295:1037. - PubMed
-
- Kuzin AM, Slozhenikina LV, Fialkovaskaia LA, Primak-Miroliubov VN. possible effect of natural background radiation on the development of mammals. Radiobiologiia. 1983;23:192–195. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

