Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae
- PMID: 19243080
- PMCID: PMC2673350
- DOI: 10.1002/yea.1658
Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae
Abstract
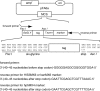
PCR-mediated gene modification is a powerful approach to the functional analysis of genes in Saccharomyces cerevisiae. One application of this method is epitope-tagging of a gene to analyse the corresponding protein by immunological methods. However, the number of epitope tags available in a convenient format is still low, and interference with protein function by the epitope, particularly if it is large, is not uncommon. To address these limitations and broaden the utility of the method, we constructed a set of convenient template plasmids designed for PCR-based C-terminal tagging with 10 different, relatively short peptide sequences that are recognized by commercially available monoclonal antibodies. The encoded tags are FLAG, 3 x FLAG, T7, His-tag, Strep-tag II, S-tag, Myc, HSV, VSV-G and V5. The same pair of primers can be used to construct tagged alleles of a gene of interest with any of the 10 tags. In addition, a six-glycine linker sequence is inserted upstream of these tags to minimize the influence of the tag on the target protein and maximize its accessibility for antibody binding. Three marker genes, HIS3MX6, kanMX6 and hphMX4, are available for each epitope. We demonstrate the utility of the new tags for both immunoblotting and one-step affinity purification of the regulatory particle of the 26S proteasome. The set of plasmids has been deposited in the non-profit plasmid repository Addgene (http://www.addgene.org).
Figures


References
-
- Ausubel FM. Current Protocols in Molecular Biology. New York: Wiley; 1987.
-
- Bahler J, Wu JQ, Longtine MS, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Brizzard B. Epitope tagging. Biotechniques. 2008;44:693–695. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

