Geometric and electronic structures of the Ni(I) and methyl-Ni(III) intermediates of methyl-coenzyme M reductase
- PMID: 19243132
- PMCID: PMC2667316
- DOI: 10.1021/bi900087w
Geometric and electronic structures of the Ni(I) and methyl-Ni(III) intermediates of methyl-coenzyme M reductase
Abstract
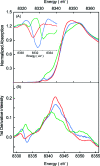
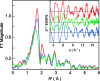
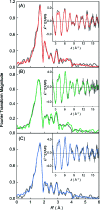
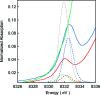
Methyl-coenzyme M reductase (MCR) catalyzes the terminal step in the formation of biological methane from methyl-coenzyme M (Me-SCoM) and coenzyme B (CoBSH). The active site in MCR contains a Ni-F(430) cofactor, which can exist in different oxidation states. The catalytic mechanism of methane formation has remained elusive despite intense spectroscopic and theoretical investigations. On the basis of spectroscopic and crystallographic data, the first step of the mechanism is proposed to involve a nucleophilic attack of the Ni(I) active state (MCR(red1)) on Me-SCoM to form a Ni(III)-methyl intermediate, while computational studies indicate that the first step involves the attack of Ni(I) on the sulfur of Me-SCoM, forming a CH(3)(*) radical and a Ni(II)-thiolate species. In this study, a combination of Ni K-edge X-ray absorption spectroscopic (XAS) studies and density functional theory (DFT) calculations have been performed on the Ni(I) (MCR(red1)), Ni(II) (MCR(red1-silent)), and Ni(III)-methyl (MCR(Me)) states of MCR to elucidate the geometric and electronic structures of the different redox states. Ni K-edge EXAFS data are used to reveal a five-coordinate active site with an open upper axial coordination site in MCR(red1). Ni K-pre-edge and EXAFS data and time-dependent DFT calculations unambiguously demonstrate the presence of a long Ni-C bond ( approximately 2.04 A) in the Ni(III)-methyl state of MCR. The formation and stability of this species support mechanism I, and the Ni-C bond length suggests a homolytic cleavage of the Ni(III)-methyl bond in the subsequent catalytic step. The XAS data provide insight into the role of the unique F(430) cofactor in tuning the stability of the different redox states of MCR.
Figures


Similar articles
-
X-ray absorption and resonance Raman studies of methyl-coenzyme M reductase indicating that ligand exchange and macrocycle reduction accompany reductive activation.J Am Chem Soc. 2002 Nov 6;124(44):13242-56. doi: 10.1021/ja020314h. J Am Chem Soc. 2002. PMID: 12405853
-
Detection of organometallic and radical intermediates in the catalytic mechanism of methyl-coenzyme M reductase using the natural substrate methyl-coenzyme M and a coenzyme B substrate analogue.Biochemistry. 2010 Dec 28;49(51):10902-11. doi: 10.1021/bi101562m. Epub 2010 Dec 2. Biochemistry. 2010. PMID: 21090696
-
Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.Biochemistry. 2010 Sep 7;49(35):7683-93. doi: 10.1021/bi100458d. Biochemistry. 2010. PMID: 20707311 Free PMC article.
-
Structural and Mechanistic Advances in the Chemistry of Methyl-Coenzyme M Reductase (MCR).Acc Chem Res. 2025 Mar 18;58(6):824-833. doi: 10.1021/acs.accounts.4c00730. Epub 2025 Mar 5. Acc Chem Res. 2025. PMID: 40042658 Review.
-
Methyl (Alkyl)-Coenzyme M Reductases: Nickel F-430-Containing Enzymes Involved in Anaerobic Methane Formation and in Anaerobic Oxidation of Methane or of Short Chain Alkanes.Biochemistry. 2019 Dec 31;58(52):5198-5220. doi: 10.1021/acs.biochem.9b00164. Epub 2019 Apr 5. Biochemistry. 2019. PMID: 30951290 Free PMC article. Review.
Cited by
-
Nickel-dependent metalloenzymes.Arch Biochem Biophys. 2014 Feb 15;544:142-52. doi: 10.1016/j.abb.2013.09.002. Epub 2013 Sep 10. Arch Biochem Biophys. 2014. PMID: 24036122 Free PMC article. Review.
-
Structure-function relationships of anaerobic gas-processing metalloenzymes.Nature. 2009 Aug 13;460(7257):814-22. doi: 10.1038/nature08299. Nature. 2009. PMID: 19675641 Review.
-
Macromolecular crystallography and biology at the Linac Coherent Light Source.J Synchrotron Radiat. 2025 May 1;32(Pt 3):548-566. doi: 10.1107/S1600577525002735. Epub 2025 Apr 23. J Synchrotron Radiat. 2025. PMID: 40266725 Free PMC article.
-
Scrutinizing metal-ligand covalency and redox non-innocence via nitrogen K-edge X-ray absorption spectroscopy.Chem Sci. 2019 Apr 17;10(19):5044-5055. doi: 10.1039/c8sc03350a. eCollection 2019 May 21. Chem Sci. 2019. PMID: 31183055 Free PMC article.
-
A Biochemical Nickel(I) State Supports Nucleophilic Alkyl Addition: A Roadmap for Methyl Reactivity in Acetyl Coenzyme A Synthase.Inorg Chem. 2019 Jul 15;58(14):8969-8982. doi: 10.1021/acs.inorgchem.8b03546. Epub 2019 Feb 21. Inorg Chem. 2019. PMID: 30788970 Free PMC article.
References
-
- Thauer R. K. (1998) Biochemistry of methanogenesis: A tribute to Marjory Stephenson. Microbiology 144, 2377–2406. - PubMed
-
- DiMarco A. A.; Bobik T. A.; Wolfe R. S. (1990) Unusual coenzymes of methanogenesis. Annu. Rev. Biochem. 59, 355–394. - PubMed
-
- Ellermann J.; Hedderich R.; Böcher R.; Thauer R. K. (1988) The final step in methane formation: Investigations with highly purified methyl-coenzyme M reductase (component C) from Methanobacterium thermoautotrophicum (strain Marburg). Eur. J. Biochem. 172, 669–678. - PubMed
-
- Färber G.; Keller W.; Kratky C.; Jaun B.; Pfaltz A.; Spinner C.; Kobelt A.; Eschenmoser A. (1991) Coenzyme F430 from methanogenic bacteria: Complete assignment of configuration based on X-ray analysis of 12,13-Diepi-F430 pentamethyl ester and on NMR spectroscopy. Helv. Chim. Acta 74, 697–716.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

