Functional correlation of bacterial LuxS with their quaternary associations: interface analysis of the structure networks
- PMID: 19243584
- PMCID: PMC2656534
- DOI: 10.1186/1472-6807-9-8
Functional correlation of bacterial LuxS with their quaternary associations: interface analysis of the structure networks
Abstract
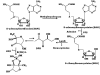
Background: The genome of a wide variety of prokaryotes contains the luxS gene homologue, which encodes for the protein S-ribosylhomocysteinelyase (LuxS). This protein is responsible for the production of the quorum sensing molecule, AI-2 and has been implicated in a variety of functions such as flagellar motility, metabolic regulation, toxin production and even in pathogenicity. A high structural similarity is present in the LuxS structures determined from a few species. In this study, we have modelled the structures from several other species and have investigated their dimer interfaces. We have attempted to correlate the interface features of LuxS with the phenotypic nature of the organisms.
Results: The protein structure networks (PSN) are constructed and graph theoretical analysis is performed on the structures obtained from X-ray crystallography and on the modelled ones. The interfaces, which are known to contain the active site, are characterized from the PSNs of these homodimeric proteins. The key features presented by the protein interfaces are investigated for the classification of the proteins in relation to their function. From our analysis, structural interface motifs are identified for each class in our dataset, which showed distinctly different pattern at the interface of LuxS for the probiotics and some extremophiles. Our analysis also reveals potential sites of mutation and geometric patterns at the interface that was not evident from conventional sequence alignment studies.
Conclusion: The structure network approach employed in this study for the analysis of dimeric interfaces in LuxS has brought out certain structural details at the side-chain interaction level, which were elusive from the conventional structure comparison methods. The results from this study provide a better understanding of the relation between the luxS gene and its functional role in the prokaryotes. This study also makes it possible to explore the potential direction towards the design of inhibitors of LuxS and thus towards a wide range of antimicrobials.
Figures








Similar articles
-
Elucidation of the conformational free energy landscape in H.pylori LuxS and its implications to catalysis.BMC Struct Biol. 2010 Aug 12;10:27. doi: 10.1186/1472-6807-10-27. BMC Struct Biol. 2010. PMID: 20704697 Free PMC article.
-
Genome-wide survey and phylogeny of S-Ribosylhomocysteinase (LuxS) enzyme in bacterial genomes.BMC Genomics. 2016 Sep 20;17(1):742. doi: 10.1186/s12864-016-3002-x. BMC Genomics. 2016. PMID: 27650568 Free PMC article.
-
A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.Structure. 2001 Jun;9(6):527-37. doi: 10.1016/s0969-2126(01)00613-x. Structure. 2001. PMID: 11435117
-
The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS.J Mol Biol. 2001 Oct 12;313(1):111-22. doi: 10.1006/jmbi.2001.5027. J Mol Biol. 2001. PMID: 11601850
-
The LuxS/AI-2 system of Streptococcus suis.Appl Microbiol Biotechnol. 2018 Sep;102(17):7231-7238. doi: 10.1007/s00253-018-9170-7. Epub 2018 Jun 25. Appl Microbiol Biotechnol. 2018. PMID: 29938319 Review.
Cited by
-
S-Ribosylhomocysteine Analogues Modified at the Ribosyl C-4 Position.J Sulphur Chem. 2016;37(3):307-327. doi: 10.1080/17415993.2015.1137921. Epub 2016 Feb 24. J Sulphur Chem. 2016. PMID: 27516805 Free PMC article.
-
Elucidation of the conformational free energy landscape in H.pylori LuxS and its implications to catalysis.BMC Struct Biol. 2010 Aug 12;10:27. doi: 10.1186/1472-6807-10-27. BMC Struct Biol. 2010. PMID: 20704697 Free PMC article.
-
Quorum sensing in thermophiles: prevalence of autoinducer-2 system.BMC Microbiol. 2018 Jun 28;18(1):62. doi: 10.1186/s12866-018-1204-x. BMC Microbiol. 2018. PMID: 29954335 Free PMC article.
References
-
- Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio Harveyi:sequence and function of genes encoding a secondary sensory pathway. Mol microbiol. 1994;13:273–286. - PubMed
-
- Zhu JDE, Hu X, Wavreille A, Park J, Pei P. S-Ribosylhomocysteinase (LuxS) Is a Mononuclear Iron Protein. Biochemistry. 2003;42:4717–4726. - PubMed
-
- De Keersmaecker SCJ, Sonck K, Vanderleyden J. Let LuxS speak up in AI-2 signaling. TRENDS in Microbiology. 2006;14:114–119. - PubMed
-
- Lewis HA, Furlong EB, Laubert B, Eroshkina GA, Batiyenko Y, Adams JM, Bergseid MG, Marsh CD, Peat TS, Sanderson WE, et al. A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs. Structure. 2001;9:527–537. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

