Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology
- PMID: 19244215
- PMCID: PMC2681385
- DOI: 10.1194/jlr.R900004-JLR200
Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology
Erratum in
- J Lipid Res. 2009 Jul;50(7):1505
Abstract
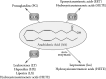
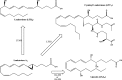
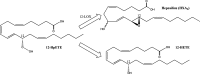
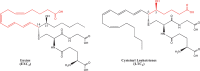
Eicosanoids have been implicated in a vast number of devastating inflammatory conditions, including arthritis, atherosclerosis, pain, and cancer. Currently, over a hundred different eicosanoids have been identified, with many having potent bioactive signaling capacity. These lipid metabolites are synthesized de novo by at least 50 unique enzymes, many of which have been cloned and characterized. Due to the extensive characterization of eicosanoid biosynthetic pathways, this field provides a unique framework for integrating genomics, proteomics, and metabolomics toward the investigation of disease pathology. To facilitate a concerted systems biology approach, this review outlines the proteins implicated in eicosanoid biosynthesis and signaling in human, mouse, and rat. Applications of the extensive genomic and lipidomic research to date illustrate the questions in eicosanoid signaling that could be uniquely addressed by a thorough analysis of the entire eicosanoid proteome.
Figures


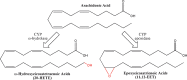

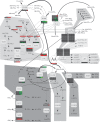
References
-
- Aderem A. 2005. Systems biology: its practice and challenges. Cell. 121 511–513. - PubMed
-
- de Godoy L. M., J. V. Olsen, J. Cox, M. L. Nielsen, N. C. Hubner, F. Frohlich, T. C. Walther, and M. Mann. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 455 1251–1254. - PubMed
-
- Funk C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294 1871–1875. - PubMed
-
- Simmons D. L., R. M. Botting, and T. Hla. 2004. Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibition. Pharmacol. Rev. 56 387–437. - PubMed
-
- Smith W. L., D. L. DeWitt, and R. M. Garavito. 2000. Cyclooxygenases: structural, cellular, and molecular biology. Annu. Rev. Biochem. 69 145–182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

