Functional complementation of UvsX and UvsY mutations in the mediation of T4 homologous recombination
- PMID: 19244311
- PMCID: PMC2673438
- DOI: 10.1093/nar/gkp096
Functional complementation of UvsX and UvsY mutations in the mediation of T4 homologous recombination
Abstract
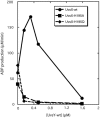
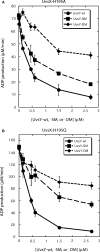
Bacteriophage T4 homologous recombination events are promoted by presynaptic filaments of UvsX recombinase bound to single-stranded DNA (ssDNA). UvsY, the phage recombination mediator protein, promotes filament assembly in a concentration-dependent manner, stimulating UvsX at stoichiometric concentrations but inhibiting at higher concentrations. Recent work demonstrated that UvsX-H195Q/A mutants exhibit decreased ssDNA-binding affinity and altered enzymatic properties. Here, we show that unlike wild-type UvsX, the ssDNA-dependent ATPase activities of UvsX-H195Q/A are strongly inhibited by both low and high concentrations of UvsY protein. This inhibition is partially relieved by UvsY mutants with decreased ssDNA-binding affinity. The UvsX-H195Q mutant retains weak DNA strand exchange activity that is inhibited by wild-type UvsY, but stimulated by ssDNA-binding compromised UvsY mutants. These and other results support a mechanism in which the formation of competent presynaptic filaments requires a hand-off of ssDNA from UvsY to UvsX, with the efficiency of the hand-off controlled by the relative ssDNA-binding affinities of the two proteins. Other results suggest that UvsY acts as a nucleotide exchange factor for UvsX, enhancing filament stability by increasing the lifetime of the high-affinity, ATP-bound form of the enzyme. Our findings reveal new details of the UvsX/UvsY relationship in T4 recombination, which may have parallels in other recombinase/mediator systems.
Figures



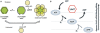
Similar articles
-
Mutations in a conserved motif inhibit single-stranded DNA binding and recombination mediator activities of bacteriophage T4 UvsY protein.J Biol Chem. 2004 Feb 13;279(7):6077-86. doi: 10.1074/jbc.M311557200. Epub 2003 Nov 22. J Biol Chem. 2004. PMID: 14634008
-
Mechanism of presynaptic filament stabilization by the bacteriophage T4 UvsY recombination mediator protein.Biochemistry. 2006 May 2;45(17):5493-502. doi: 10.1021/bi0525167. Biochemistry. 2006. PMID: 16634631
-
Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions.J Biol Chem. 2006 Sep 8;281(36):26308-19. doi: 10.1074/jbc.M604349200. Epub 2006 Jul 7. J Biol Chem. 2006. PMID: 16829679
-
Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8298-305. doi: 10.1073/pnas.131007498. Proc Natl Acad Sci U S A. 2001. PMID: 11459967 Free PMC article. Review.
-
DNA strand exchange proteins: a biochemical and physical comparison.Front Biosci. 1998 Jun 17;3:D570-603. doi: 10.2741/a304. Front Biosci. 1998. PMID: 9632377 Review.
Cited by
-
An archaeal RadA paralog influences presynaptic filament formation.DNA Repair (Amst). 2013 Jun 1;12(6):403-13. doi: 10.1016/j.dnarep.2013.03.003. Epub 2013 Apr 24. DNA Repair (Amst). 2013. PMID: 23622866 Free PMC article.
-
Bacteriophage T4 Escapes CRISPR Attack by Minihomology Recombination and Repair.mBio. 2021 Jun 29;12(3):e0136121. doi: 10.1128/mBio.01361-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154416 Free PMC article.
-
Assembly and dynamics of the bacteriophage T4 homologous recombination machinery.Virol J. 2010 Dec 3;7:357. doi: 10.1186/1743-422X-7-357. Virol J. 2010. PMID: 21129202 Free PMC article. Review.
-
Presynaptic filament dynamics in homologous recombination and DNA repair.Crit Rev Biochem Mol Biol. 2011 Jun;46(3):240-70. doi: 10.3109/10409238.2011.576007. Crit Rev Biochem Mol Biol. 2011. PMID: 21599536 Free PMC article.
-
DNA-binding properties of T4 UvsY recombination mediator protein: polynucleotide wrapping promotes high-affinity binding to single-stranded DNA.Nucleic Acids Res. 2010 Aug;38(14):4821-33. doi: 10.1093/nar/gkq219. Epub 2010 Apr 5. Nucleic Acids Res. 2010. PMID: 20371513 Free PMC article.
References
-
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. - PubMed
-
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. - PubMed
-
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. - PubMed
-
- Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–8993. - PubMed

