Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection
- PMID: 19244322
- PMCID: PMC2668508
- DOI: 10.1128/JVI.00042-09
Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection
Abstract
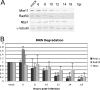
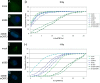
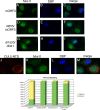
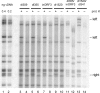
Adenovirus infection induces a cellular DNA damage response that can inhibit viral DNA replication and ligate viral genomes into concatemers. It is not clear if the input virus is sufficient to trigger this response or if viral DNA replication is required. Adenovirus has evolved two mechanisms that target the Mre11-Rad50-Nbs1 (MRN) complex to inhibit the DNA damage response. These include E4-ORF3-dependent relocalization of MRN proteins and E4-ORF6/E1B-55K-dependent degradation of MRN components. The literature suggests that degradation of the MRN complex due to E4-ORF6/E1B-55K does not occur until after viral DNA replication has begun. We show that, by the time viral DNA accumulates, the MRN complex is inactivated by either of the E4-induced mechanisms and that, with E4-ORF6/E1B-55K, this inactivation is due to MRN degradation. Our data are consistent with the conclusion that input viral DNA is sufficient to induce the DNA damage response. Further, we demonstrate that when the DNA damage response is active in E4 mutant virus infections, the covalently attached terminal protein is not cleaved from viral DNAs, and the viral origins of replication are not detectably degraded at a time corresponding to the onset of viral replication. The sequences of concatemeric junctions of viral DNAs were determined, which supports the conclusion that nonhomologous end joining mediates viral DNA ligation. Large deletions were found at these junctions, demonstrating nucleolytic procession of the viral DNA; however, the lack of terminal protein cleavage and terminus degradation at earlier times shows that viral genome deletion and concatenation are late effects.
Figures

Similar articles
-
Adenovirus regulates sumoylation of Mre11-Rad50-Nbs1 components through a paralog-specific mechanism.J Virol. 2012 Sep;86(18):9656-65. doi: 10.1128/JVI.01273-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740413 Free PMC article.
-
Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication.J Virol. 2005 May;79(10):6207-15. doi: 10.1128/JVI.79.10.6207-6215.2005. J Virol. 2005. PMID: 15858005 Free PMC article.
-
Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation.J Virol. 2007 Jul;81(13):7034-40. doi: 10.1128/JVI.00029-07. Epub 2007 Apr 25. J Virol. 2007. PMID: 17459921 Free PMC article.
-
MRN complex is an essential effector of DNA damage repair.J Zhejiang Univ Sci B. 2021 Jan 15;22(1):31-37. doi: 10.1631/jzus.B2000289. J Zhejiang Univ Sci B. 2021. PMID: 33448185 Free PMC article. Review.
-
Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair.Prog Biophys Mol Biol. 2015 Mar;117(2-3):182-193. doi: 10.1016/j.pbiomolbio.2014.12.004. Epub 2015 Jan 7. Prog Biophys Mol Biol. 2015. PMID: 25576492 Free PMC article. Review.
Cited by
-
Differential activation of cellular DNA damage responses by replication-defective and replication-competent adenovirus mutants.J Virol. 2012 Dec;86(24):13324-33. doi: 10.1128/JVI.01757-12. Epub 2012 Sep 26. J Virol. 2012. PMID: 23015708 Free PMC article.
-
Viral DNA replication-dependent DNA damage response activation during BK polyomavirus infection.J Virol. 2015 May;89(9):5032-9. doi: 10.1128/JVI.03650-14. Epub 2015 Feb 18. J Virol. 2015. PMID: 25694603 Free PMC article.
-
The α2 helix in the DNA ligase IV BRCT-1 domain is required for targeted degradation of ligase IV during adenovirus infection.Virology. 2012 Jul 5;428(2):128-35. doi: 10.1016/j.virol.2012.03.006. Epub 2012 Apr 24. Virology. 2012. PMID: 22534089 Free PMC article.
-
Impact of Adenovirus E4-ORF3 Oligomerization and Protein Localization on Cellular Gene Expression.Viruses. 2015 May 13;7(5):2428-49. doi: 10.3390/v7052428. Viruses. 2015. PMID: 25984715 Free PMC article.
-
Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies.J Virol. 2010 Dec;84(23):12210-25. doi: 10.1128/JVI.01442-10. Epub 2010 Sep 22. J Virol. 2010. PMID: 20861261 Free PMC article.
References
-
- Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421499-506. - PubMed
-
- Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156107-121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

