The origins of phagocytosis and eukaryogenesis
- PMID: 19245710
- PMCID: PMC2651865
- DOI: 10.1186/1745-6150-4-9
The origins of phagocytosis and eukaryogenesis
Abstract
Background: Phagocytosis, that is, engulfment of large particles by eukaryotic cells, is found in diverse organisms and is often thought to be central to the very origin of the eukaryotic cell, in particular, for the acquisition of bacterial endosymbionts including the ancestor of the mitochondrion.
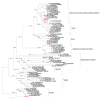
Results: Comparisons of the sets of proteins implicated in phagocytosis in different eukaryotes reveal extreme diversity, with very few highly conserved components that typically do not possess readily identifiable prokaryotic homologs. Nevertheless, phylogenetic analysis of those proteins for which such homologs do exist yields clues to the possible origin of phagocytosis. The central finding is that a subset of archaea encode actins that are not only monophyletic with eukaryotic actins but also share unique structural features with actin-related proteins (Arp) 2 and 3. All phagocytic processes are strictly dependent on remodeling of the actin cytoskeleton and the formation of branched filaments for which Arp2/3 are responsible. The presence of common structural features in Arp2/3 and the archaeal actins suggests that the common ancestors of the archaeal and eukaryotic actins were capable of forming branched filaments, like modern Arp2/3. The Rho family GTPases that are ubiquitous regulators of phagocytosis in eukaryotes appear to be of bacterial origin, so assuming that the host of the mitochondrial endosymbiont was an archaeon, the genes for these GTPases come via horizontal gene transfer from the endosymbiont or in an earlier event.
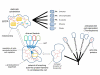
Conclusion: The present findings suggest a hypothetical scenario of eukaryogenesis under which the archaeal ancestor of eukaryotes had no cell wall (like modern Thermoplasma) but had an actin-based cytoskeleton including branched actin filaments that allowed this organism to produce actin-supported membrane protrusions. These protrusions would facilitate accidental, occasional engulfment of bacteria, one of which eventually became the mitochondrion. The acquisition of the endosymbiont triggered eukaryogenesis, in particular, the emergence of the endomembrane system that eventually led to the evolution of modern-type phagocytosis, independently in several eukaryotic lineages.
Figures


References
-
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. - PubMed
-
- Esser C, Ahmadinejad N, Wiegand C, Rotte C, Sebastiani F, Gelius-Dietrich G, Henze K, Kretschmann E, Richly E, Leister D, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–1660. - PubMed
-
- Gray MW. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:233–357. - PubMed
-
- Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. - PubMed
-
- Roger AJ. Reconstructing Early Events in Eukaryotic Evolution. Am Nat. 1999;154:S146–S163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

