Little but loud: small RNAs have a resounding affect on ear development
- PMID: 19245798
- PMCID: PMC2700218
- DOI: 10.1016/j.brainres.2009.02.027
Little but loud: small RNAs have a resounding affect on ear development
Abstract
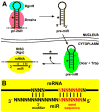
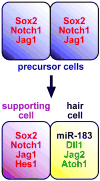
The impact of small RNA function has resonated throughout nearly every aspect of eukaryotic biology and captured the varied interests of researchers, whether they are endeavoring to understand the basis of development and disease or seeking novel therapeutic targets and tools. The genetic regulatory roles of microRNAs (miRNAs) are particularly interesting given that these often highly conserved factors post-transcriptionally silence many complementary target genes by inhibiting messenger RNA translation. In this regard, miRNAs can be considered as counterparts to transcription factors, the ensemble of which establishes the set of expressed genes that define the characteristics of a specific cell type. In this review, evidence supporting a resounding role for small RNAs in development and maturation of sensory epithelia in the mouse inner ear will be considered with an emphasis on the contribution of one hair cell miRNA family (miR-183, miR-96, and miR-182). Although there is much yet to be explored in this fledgling aspect of ear biology, the breadth of miRNA expression and functional requirement for ear development are already sounding off.
Figures
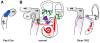

Similar articles
-
MicroRNA gene expression in the mouse inner ear.Brain Res. 2006 Sep 21;1111(1):95-104. doi: 10.1016/j.brainres.2006.07.006. Epub 2006 Aug 10. Brain Res. 2006. PMID: 16904081
-
Next-generation sequencing of small RNAs from inner ear sensory epithelium identifies microRNAs and defines regulatory pathways.BMC Genomics. 2014 Jun 18;15(1):484. doi: 10.1186/1471-2164-15-484. BMC Genomics. 2014. PMID: 24942165 Free PMC article.
-
Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear.Gene Expr Patterns. 2009 Jun;9(5):364-70. doi: 10.1016/j.gep.2009.01.003. Gene Expr Patterns. 2009. PMID: 19602392
-
MicroRNAs and epigenetic regulation in the mammalian inner ear: implications for deafness.Mamm Genome. 2009 Sep-Oct;20(9-10):581-603. doi: 10.1007/s00335-009-9230-5. Epub 2009 Oct 30. Mamm Genome. 2009. PMID: 19876605 Review.
-
Unraveling inner ear induction by gene manipulation using Pax2-Cre BAC transgenic mice.Brain Res. 2009 Jun 24;1277:84-9. doi: 10.1016/j.brainres.2009.02.036. Epub 2009 Mar 2. Brain Res. 2009. PMID: 19265685 Review.
Cited by
-
Circulating Serum miRNA-205 as a Diagnostic Biomarker for Ototoxicity in Mice Treated with Aminoglycoside Antibiotics.Int J Mol Sci. 2018 Sep 19;19(9):2836. doi: 10.3390/ijms19092836. Int J Mol Sci. 2018. PMID: 30235835 Free PMC article.
-
The genetics of hair cell development and regeneration.Annu Rev Neurosci. 2013 Jul 8;36:361-81. doi: 10.1146/annurev-neuro-062012-170309. Epub 2013 May 29. Annu Rev Neurosci. 2013. PMID: 23724999 Free PMC article. Review.
-
Evolution of microRNAs and the diversification of species.Genome Biol Evol. 2011;3:55-65. doi: 10.1093/gbe/evq085. Epub 2010 Dec 15. Genome Biol Evol. 2011. PMID: 21169229 Free PMC article.
-
The microRNA-183 cluster: the family that plays together stays together.Nucleic Acids Res. 2015 Sep 3;43(15):7173-88. doi: 10.1093/nar/gkv703. Epub 2015 Jul 13. Nucleic Acids Res. 2015. PMID: 26170234 Free PMC article. Review.
-
MicroRNAs in hair cell development and deafness.Curr Opin Otolaryngol Head Neck Surg. 2010 Oct;18(5):459-65. doi: 10.1097/MOO.0b013e32833e0601. Curr Opin Otolaryngol Head Neck Surg. 2010. PMID: 20717030 Free PMC article.
References
-
- Agirre X, Jiménez-Velasco A, San José-Enériz E, Garate L, Bandrés E, Cordeu L, Aparicio O, Saez B, Navarro G, Vilas-Zornoza A, Pérez-Roger I, García-Foncillas J, Torres A, Heiniger A, Calasanz MJ, Fortes P, Román-Gómez J, Prósper F. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res. 2008;6:1830–1840. - PubMed
-
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome is an RNA machine. Science. 2008;319:1787–1789. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

