DNA-lesion mapping in mammalian cells
- PMID: 19245834
- PMCID: PMC2693248
- DOI: 10.1016/j.ymeth.2009.02.008
DNA-lesion mapping in mammalian cells
Abstract
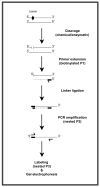
Formation of DNA damage is a crucial event in carcinogenesis. Irreparable DNA lesions have the potential to cause mispairing during DNA replication, thereby giving rise to mutations. Critically important mutations in cancer-related genes, i.e., oncogenes and tumor suppressor genes, are key contributors to carcinogenesis. Theoretically, co-localization(s) of persistent DNA lesions and mutational hotspots in cancer-relevant genes can be used for causality inference. The inferred causality can be validated if a suspected carcinogen can similarly produce corresponding patterns of DNA damage and mutagenesis in vitro and/or in vivo. DNA-lesion footprinting (mapping) in conjunction with mutagenicity analysis is used for investigating cancer etiology. Ligation-mediated polymerase chain reaction (LM-PCR) is a versatile DNA-lesion footprinting technique, which enables sensitive and specific detection of DNA damage, at the level of nucleotide resolution, in genomic DNA. Here, we describe an updated protocol for LM-PCR analysis of the mammalian genome. This protocol can routinely be used for DNA-lesion footprinting of a variety of chemical and/or physical carcinogens in mammalian cells.
Figures


References
-
- Besaratinia A, Pfeifer GP. Carcinogenesis. 2006;27:1526–37. - PubMed
-
- Denissenko MF, Pao A, Tang M, Pfeifer GP. Science. 1996;274:430–32. - PubMed
-
- Chen JX, Zheng Y, West M, Tang MS. Cancer Res. 1998;58:2070–75. - PubMed
-
- DeMarini DM, Landi S, Tian D, Hanley NM, Li X, Hu F, Roop BC, Mass MJ, Keohavong P, Gao W, Olivier M, Hainaut P, Mumford JL. Cancer Res. 2001;61:6679–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

