Structure, expression, and biological function of INSM1 transcription factor in neuroendocrine differentiation
- PMID: 19246490
- PMCID: PMC2704596
- DOI: 10.1096/fj.08-125971
Structure, expression, and biological function of INSM1 transcription factor in neuroendocrine differentiation
Abstract
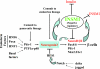
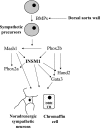
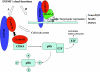
Zinc-finger transcription factors are DNA-binding proteins that are implicated in many diverse biological functions. INSM1 (formerly IA-1) contains five zinc-finger motifs and functions as a transcription factor. INSM1 protein structure is highly conserved in homologues of different species. It is predominantly expressed in developing neuroendocrine tissues and the nervous system in mammals. INSM1 represents an important player in early embryonic neurogenesis. In pancreatic endocrine cell differentiation, Ngn3 first activates INSM1 and subsequently NeuroD/beta2. Conversely, INSM1 exerts a feedback mechanism to suppress NeuroD/beta2 and its own gene expression. INSM1 gene ablation in the mouse results in the impairment of pancreatic endocrine cell maturation. Further, deletion of INSM1 severely impairs catecholamine biosynthesis and secretion from the adrenal gland that results in early embryonic lethality. Genetically, INSM1 acts as a downstream factor of Mash 1 and Phox2b in the differentiation of the sympatho-adrenal lineage. In the developing neocortex, mouse embryos lacking INSM1 expression contain half the number of basal progenitors and show a reduction in cortical plate radial thickness. Cell signaling studies reveal that INSM1 contributes to the induction of cell cycle arrest/exit necessary to facilitate cellular differentiation. INSM1 is highly expressed in tumors of neuroendocrine origin. Hence, its promoter could serve as a tumor-specific promoter that drives a specific targeted cancer gene therapy for the treatment of neuroendocrine tumors. Taken together, all of these features of INSM1 strongly support its role as an important regulator during neuroendocrine differentiation.
Figures


References
-
- Goto Y, DeSilva M G, Toscani A, Prabhakar B S, Notkins A L, Lan M S. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with zinc-finger DNA-binding motifs. J Biol Chem. 1992;267:15252–15257. - PubMed
-
- Leslie R D G, Atkinson M A, Notkins A L. Autoantigens IA-2 and GAD in type 1 (insulin-dependent) diabetes. Diabetologia. 1999;42:3–14. - PubMed
-
- Lan M S, Li Q, Lu J, Modi W S, Notkins A L. Genomic organization, 5′-upstream sequence, and chromosomal localization of an insulinoma-associated intronless gene, IA-1. J Biol Chem. 1994;269:14170–14174. - PubMed
-
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

