Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia
- PMID: 19246560
- PMCID: PMC2686141
- DOI: 10.1182/blood-2008-11-189845
Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia
Abstract
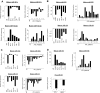
Human T-cell leukemia virus type-I (HTLV-I) is the etiologic agent of adult T-cell leukemia (ATL), an aggressive lymphoproliferative disease. MicroRNAs (miRNAs) are differentially expressed during hematopoiesis and lineage commitment of hematopoietic stem cell progenitors (HSCPs). Here, we report aberrant expression of hematopoietic-specific miR-223, miR-181a, miR-150, miR-142.3p, and miR-155 in HTLV-I-infected cells in vitro and uncultured ex vivo ATL cells. Our results suggest that HTLV-I-infected cells have an unbalanced expression of miRNA that favors T-cell differentiation. We also found altered expression of miRNA previously recognized as innate immunity regulators: miR-155, miR-125a, miR-132, and miR-146. Strikingly, our data also revealed significant differences between ex vivo ATL tumor cells and in vitro HTLV-I cell lines. Specifically, miR-150 and miR-223 were up-regulated in ATL patients but consistently down-regulated in HTLV-I cell lines, suggesting that ATL cells and in vitro-established cells are derived from distinct cellular populations.
Figures
References
-
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. - PubMed
-
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. - PubMed
-
- Kluiver J, Kroesen BJ, Poppema S, van den BA. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20:1931–1936. - PubMed
-
- García P, Frampton J. Hematopoietic lineage commitment: miRNAs add specificity to a widely expressed transcription factor. Dev Cell. 2008;14:815–816. - PubMed
-
- Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

