Preventing oxidative stress: a new role for XBP1
- PMID: 19247368
- PMCID: PMC2826168
- DOI: 10.1038/cdd.2009.14
Preventing oxidative stress: a new role for XBP1
Abstract
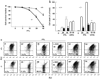
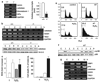
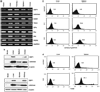
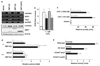
Antioxidant molecules reduce oxidative stress and protect cells from reactive oxygen species (ROS)-mediated cellular damage and probably the development of cancer. We have investigated the contribution of X-box-binding protein (XBP1), a major endoplasmic reticulum stress-linked transcriptional factor, to cellular resistance to oxidative stress. After exposure to hydrogen peroxide (H(2)O(2)) or a strong ROS inducer parthenolide, loss of mitochondrial membrane potential (MMP) and subsequent cell death occurred more extensively in XBP1-deficient cells than wild-type mouse embryonic fibroblast cells, whereas two other anticancer agents induced death similarly in both cells. In XBP1-deficient cells, H(2)O(2) exposure induced more extensive ROS generation and prolonged p38 phosphorylation, and expression of several antioxidant molecules including catalase was lower. Knockdown of XBP1 decreased catalase expression, enhanced ROS generation and MMP loss after H(2)O(2) exposure, but extrinsic catalase supply rescued them. Overexpression of XBP1 recovered catalase expression in XBP1-deficient cells and diminished ROS generation after H(2)O(2) exposure. Mutation analysis of the catalase promoter region suggests a pivotal role of CCAAT boxes, NF-Y-binding sites, for the XBP1-mediated enhancing effect. Taken together, these results indicate a protective role of XBP1 against oxidative stress, and its positive regulation of catalase expression may at least in part account for this function.
Figures



Similar articles
-
Targeting X box-binding protein-1 (XBP1) enhances sensitivity of glioma cells to oxidative stress.Neuropathol Appl Neurobiol. 2011 Jun;37(4):395-405. doi: 10.1111/j.1365-2990.2010.01155.x. Neuropathol Appl Neurobiol. 2011. PMID: 21138464
-
Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II.J Biochem. 2004 Sep;136(3):343-50. doi: 10.1093/jb/mvh122. J Biochem. 2004. PMID: 15598891
-
Preconditioning with endoplasmic reticulum stress mitigates retinal endothelial inflammation via activation of X-box binding protein 1.J Biol Chem. 2011 Feb 11;286(6):4912-21. doi: 10.1074/jbc.M110.199729. Epub 2010 Dec 7. J Biol Chem. 2011. PMID: 21138840 Free PMC article.
-
Hepatocyte X-box binding protein 1 deficiency increases liver injury in mice fed a high-fat/sugar diet.Am J Physiol Gastrointest Liver Physiol. 2015 Dec 15;309(12):G965-74. doi: 10.1152/ajpgi.00132.2015. Epub 2015 Oct 15. Am J Physiol Gastrointest Liver Physiol. 2015. PMID: 26472223 Free PMC article.
-
Activation of the UPR protects against cigarette smoke-induced RPE apoptosis through up-regulation of Nrf2.J Biol Chem. 2015 Feb 27;290(9):5367-80. doi: 10.1074/jbc.M114.603738. Epub 2015 Jan 7. J Biol Chem. 2015. PMID: 25568320 Free PMC article.
Cited by
-
Endoplasmic reticulum stress signalling - from basic mechanisms to clinical applications.FEBS J. 2019 Jan;286(2):241-278. doi: 10.1111/febs.14608. Epub 2018 Aug 4. FEBS J. 2019. PMID: 30027602 Free PMC article. Review.
-
The Endoplasmic Reticulum Unfolded Protein Response in Neurodegenerative Disorders and Its Potential Therapeutic Significance.Front Mol Neurosci. 2017 Jun 16;10:187. doi: 10.3389/fnmol.2017.00187. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28670265 Free PMC article. Review.
-
Multi-System-Level Analysis with RNA-Seq on Pterygium Inflammation Discovers Association between Inflammatory Responses, Oxidative Stress, and Oxidative Phosphorylation.Int J Mol Sci. 2024 Apr 27;25(9):4789. doi: 10.3390/ijms25094789. Int J Mol Sci. 2024. PMID: 38732006 Free PMC article.
-
The unfolded protein response and diabetic retinopathy.J Diabetes Res. 2014;2014:160140. doi: 10.1155/2014/160140. Epub 2014 Oct 29. J Diabetes Res. 2014. PMID: 25530974 Free PMC article. Review.
-
TXNIP links innate host defense mechanisms to oxidative stress and inflammation in retinal Muller glia under chronic hyperglycemia: implications for diabetic retinopathy.Exp Diabetes Res. 2012;2012:438238. doi: 10.1155/2012/438238. Epub 2012 Mar 18. Exp Diabetes Res. 2012. PMID: 22474421 Free PMC article.
References
-
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. - PubMed
-
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. - PubMed
-
- Deisseroth A, Dounce AL. Catalase: Physical and chemical properties, mechanism of catalysis, physiological role. Physiol Rev. 1970;50:319–375. - PubMed
-
- Schisler NJ, Singh SM. Inheritance and expression of tissue-specific catalase activity during development and aging in mice. Genome. 1987;29:748–760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

