A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors
- PMID: 19247443
- PMCID: PMC2642726
- DOI: 10.1371/journal.pgen.1000396
A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors
Abstract
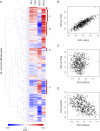
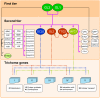
Position-dependent cell fate determination and pattern formation are unique aspects of the development of plant structures. The establishment of single-celled leaf hairs (trichomes) from pluripotent epidermal (protodermal) cells in Arabidopsis provides a powerful system to determine the gene regulatory networks involved in cell fate determination. To obtain a holistic view of the regulatory events associated with the differentiation of Arabidopsis epidermal cells into trichomes, we combined expression and genome-wide location analyses (ChIP-chip) on the trichome developmental selectors GLABRA3 (GL3) and GLABRA1 (GL1), encoding basic helix-loop-helix (bHLH) and MYB transcription factors, respectively. Meta-analysis was used to integrate genome-wide expression results contrasting wild type and gl3 or gl1 mutants with changes in gene expression over time using inducible versions of GL3 and GL1. This resulted in the identification of a minimal set of genes associated with the differentiation of epidermal cells into trichomes. ChIP-chip experiments, complemented by the targeted examination of factors known to participate in trichome initiation or patterning, identified about 20 novel GL3/GL1 direct targets. In addition to genes involved in the control of gene expression, such as the transcription factors SCL8 and MYC1, we identified SIM (SIAMESE), encoding a cyclin-dependent kinase inhibitor, and RBR1 (RETINOBLASTOMA RELATED1), corresponding to a negative regulator of the cell cycle transcription factor E2F, as GL3/GL1 immediate targets, directly implicating these trichome regulators in the control of the endocycle. The expression of many of the identified GL3/GL1 direct targets was specific to very early stages of trichome initiation, suggesting that they participate in some of the earliest known processes associated with protodermal cell differentiation. By combining this knowledge with the analysis of genes associated with trichome formation, our results reveal the architecture of the top tiers of the hierarchical structure of the regulatory network involved in epidermal cell differentiation and trichome formation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Serna L, Martin C. Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 2006;11:274–280. - PubMed
-
- Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu Rev Plant Biol. 2003;54:403–430. - PubMed
-
- Schellmann S, Hulskamp M. Epidermal differentiation: trichomes in Arabidopsis as a model system. Int J Dev Biol. 2005:579–584. - PubMed
-
- Szymanski D, Lloyd A, Marks MD. Progress in the molecular genetic analysis of trichome initiation and morhpogenesis in Arabidopsis. Trends Plant Sci. 2000;5:214–219. - PubMed
-
- Hulskamp M, Misera S, Jurgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

