Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates
- PMID: 19250912
- PMCID: PMC2669153
- DOI: 10.1016/j.molcel.2009.01.021
Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates
Abstract
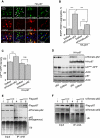
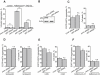
The two main routes that cells use for degrading intracellular proteins are the ubiquitin-proteasome and autophagy-lysosome pathways, which have been thought to have largely distinct clients. Here, we show that autophagy inhibition increases levels of proteasome substrates. This is largely due to p62 (also called A170/SQSTM1) accumulation after autophagy inhibition. Excess p62 inhibits the clearance of ubiquitinated proteins destined for proteasomal degradation by delaying their delivery to the proteasome's proteases. Our data show that autophagy inhibition, which was previously believed to only affect long-lived proteins, will also compromise the ubiquitin-proteasome system. This will lead to increased levels of short-lived regulatory proteins, like p53, as well as the accumulation of aggregation-prone proteins, with predicted deleterious consequences.
Figures

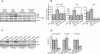



References
-
- Bennett E.J., Shaler T.A., Woodman B., Ryu K.-Y., Zaitseva T.S., Becker C.H., Bates G.P., Schulman H., Kopito R.R. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–708. - PubMed
-
- Dantuma N.P., Lindsten K., Glas R., Jellne M., Masucci M.G. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 2000;18:538–543. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

