The structure and function of insulin: decoding the TR transition
- PMID: 19251033
- PMCID: PMC3297421
- DOI: 10.1016/S0083-6729(08)00602-X
The structure and function of insulin: decoding the TR transition
Abstract
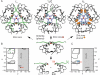
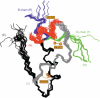
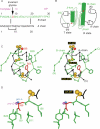
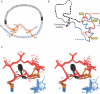
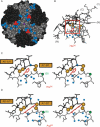
Crystal structures of insulin are remarkable for a long-range reorganization among three families of hexamers (designated T(6), T(3)R(3)(f), and R(6)). Although these structures are well characterized at atomic resolution, the biological implications of the TR transition remain the subject of speculation. Recent studies indicate that such allostery reflects a structural switch between distinct folding-competent and active conformations. Stereospecific modulation of this switch by corresponding d- and l-amino-acid substitutions yields reciprocal effects on protein stability and receptor-binding activity. Naturally occurring human mutations at the site of conformational change impair the folding of proinsulin and cause permanent neonatal-onset diabetes mellitus. The repertoire of classical structures thus foreshadows the conformational lifecycle of insulin in vivo. By highlighting the richness of information provided by protein crystallography-even in a biological realm far removed from conditions of crystallization-these findings validate the prescient insights of the late D. C. Hodgkin. Future studies of the receptor-bound structure of insulin may enable design of novel agonists for the treatment of diabetes mellitus.
Figures

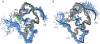
References
-
- Adams MJ, Blundell TL, Dodson EJ, Dodson GG, Vijayan M, Baker EN, Hardine MM, Hodgkin DC, Rimer B, Sheet S. Structure of rhombohedral 2 zinc insulin crystals. Nature. 1969;224:491–495.
-
- Araki E, Oyadomari S, Mori M. Endoplasmic reticulum stress and diabetes mellitus. Intern. Med. 2003;42:7–14. - PubMed
-
- Baker EN, Blundell TL, Cutfield JF, Cutfield SM, Dodson EJ, Dodson GG, Hodgkin DM, Hubbard RE, Isaacs NW, Reynolds CD. The structure of 2Zn pig insulin crystals at 1.5Å resolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1988;319:369–456. - PubMed
-
- Bentley G, Dodson E, Dodson G, Hodgkin D, Mercola D. Structure of insulin in 4-zinc insulin. Nature. 1976;261:166–168. - PubMed
-
- Bentley G, Dodson G, Lewitova A. Rhombohedral insulin crystal transformation. J. Mol. Biol. 1978;126:871–875. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

